Author
Title
Subtitle
Year
Setting
Objective
Methods
Outcome
Results
Conclusion
Evaluation
Miles, Simon; Groth, Paul; Munroe, Steve; Moreau, Luc
PrIMe
A Methodology for Developing Provenance-Aware Applications
2011
The work described in this article was undertaken in the context of a major research
investigation on provenance, which resulted in a novel approach to provenance that
it is not constrained to a single technology or application domain [...]
This article extends and gives a more detailed analysis of a preliminary discussion
of PrIMe presented at the Sixth International Software Engineering and Middleware
Workshop 2006 [Munroe et al. 2006].
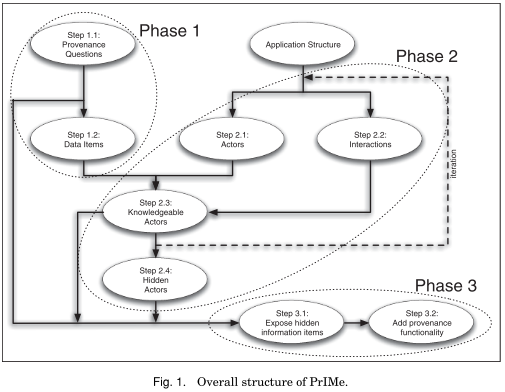
This article presents PrIMe, a software engineering technique for making applications
provenance-aware, that is, for identifying what documentation is needed to answer
provenance questions, analyzing application designs to determine at what points
process documentation will be recorded, and adapting application designs to produce
process documentation.
[Unspecified. Natural-language definitions are used, non-standard diagrams and pseudo-code algorithms shown.]
“Definition 1. Provenance of a Piece of Data. The provenance of a data item is the process that led to that data item.”
In Section 4, an example application from
bioinformatics is introduced in order to ground the subsequent discussion and explanation.
The data model in which process documentation is stored is specified in Section 5.
In Section 6, the process for identifying provenance questions is described. Section 7
then describes how to decompose applications and how to map out the flow of information
within them. Section 8 goes on to describe different kinds of adaptations that can
be made to applications in order to facilitate the recording of process documentation. In
order to assess the benefit of our new approach, we perform a comparison with an existing,
aspect-oriented approach in Section 9 [...]
[A method (descriptive);
description of processes, methods, structures, an architecture;
use case based evaluation: “what?”]
Full results omitted; see full text.
As shown in our comparison with an aspect-oriented methodology, PrIMe crystallizes issues that will arise in any project to build a provenance-aware system.
Almeida, Fernanda Nascimento; Tunes, Gisela; Brettas da Costa, Julio Cezar; Sabino, Ester Cerdeira; Mendrone-Junior, Alfredo; Ferreira, Joao Eduardo
A provenance model based on declarative specifications for intensive data analyses in hemotherapy information systems
2016
This paper is an interdisciplinary effort involving researchers from the university of São Paulo and the São Paulo Blood Center, the largest blood center in latin america.
The goal is to improve the data quality of a blood donation database, maintained by the São Paulo Blood Center, with the help of a provenance model. Better data quality, in turn, should allow to answer two main questions -- (i) ‘‘which blood donor population can be considered under risk of developing iron deficiency anemia due to multiple blood donations?’’, and (ii) ‘‘what is the number of donations or the donation interval that increases the chances of the donor developing anemia?’’ -- and reduce undesirable outcomes for blood donors.
the provenance description was adopted for main two reasons: (i) to provide clear and precise information to the researchers regarding the data transformation process; and (ii) to allow control of each step of the data processing. This enables the re-execution of the experiments and activities when necessary. Another equally important aspect is to ensure that each process of data storing contributes to the audition. Our intention in this paper was to use the provenance description to assist in the development of data classification criteria; therefore, we used the semantic concepts underlying the questions.
[Unspecified.]
The authors mention a definition of provenance as
“the documentation of the history of the data. This documentation contains each step of the data transformation process of the data source.”
Methods:
- we established actions to quantify data for each attribute to verify those that were more suitable for analysis (resulting in Tab. 1):
- we defined Useful Data for each attribute as the fraction of valid entries for that attribute
- we rank the attributes according to their subjectivity. Therefore, we created a measurement called Reliability. Attributes that were solely declared by the subject and not verified were considered highly subjective
- The criteria for classification were based on useful data greater or equal to 80% and ‘high’ reliability; data that met these criteria were selected and considered variables of interest. It is important to take into account that only reliable variables with higher quality registers were used for the analyses. We also discarded donors who were under 18 or above 69 years of age.
- we implemented routines for treatment, cleaning, normalization, and data extraction
- Because [the main questions] cannot be answered by a simple database search, we fragmented these declarations and called them inclusion criteria.
- (preliminary) descriptive statistical analysis based on mean plots and a life table estimator [26] and multivariate analysis based on a logistic mixed model
- We sought to investigate which groups of donors had a higher chance of developing anemia using information from the previous donation. A logistic model is appropriate because our response variable is binary (anemic/non-anemic). However, because we have several donations made by the same donor in our data, a mixed model must be used to take into account the possible dependence among donations made by the same donor [28].
- Due to the differences between male and female donors, the analysis was performed separately for each group.
- We included the following variables for both analyses: age at previous donation (<50 year/50 years and over), Hct level at previous donation and time elapsed since last donation (up to 4 months/>4 months and less or equal to 8 months/more than 8 months). Different categories for Hct levels were used for males and females. Likelihood ratio tests were performed to measure the variance of the random effects, (Most of the Brazilian Blood Centers use Hct levels to prevent donations from donor with risks, i.e. no verification of ferritin levels, but correlation between the two has been shown)
An approach based on questions designed to understand the reasons for deferrals due to LHct in regular blood donors.
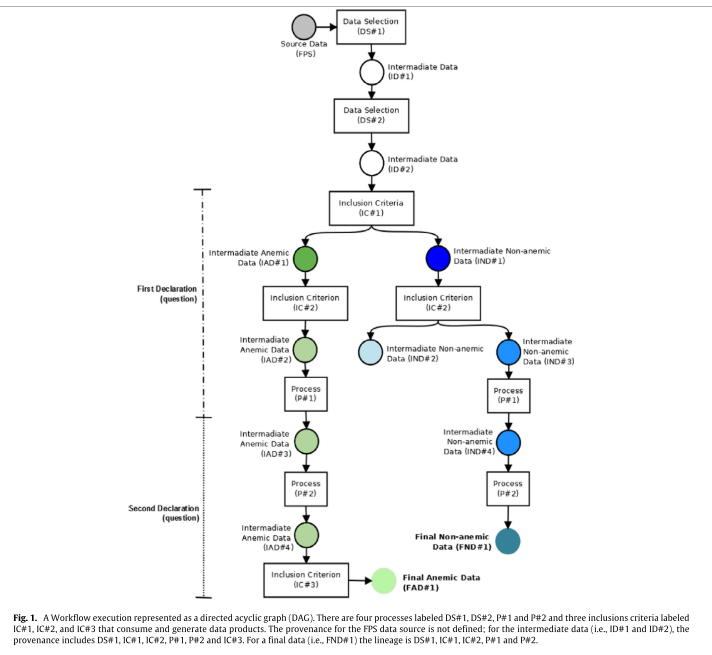
The provenance process summarized in Fig. 1 shows the set of rules (inclusion criteria) and processes used to obtain the study groups for this paper. The transformation processes are modeled as filters, and their routines and their data products serve as inputs and outputs for these tasks. The first process (DS#1, in Fig. 1) was used to select records for the period studied (from 1996 to 2006). The second process (DS# 2, in Fig. 1) is the dataset cleaning step that is composed of a set of routines that have data cleaning purposes related to the variables of interest.
- The first inclusion criterion (IC#1, Fig. 1) was implemented to classify and separate blood donors according to their risk of developing anemia. To classify these donors we used the Brazilian National Health Surveillance Agency cut-off values for Hb and Hct ... Thus, we obtained two groups of donors: anemic and non-anemic (IAD#1 and IND#1, respectively, in Fig. 1).
- For the second inclusion criterion (IC#2, in Fig. 1), we separated the donor groups according to the number ofdonations performed. We obtained three subgroups of donors that were defined as: (i) non-anemic first-time donors (IND#2), or those who had donated only once; (ii) non-anemic donors (IND#3 and IND#4), or people who donated two or more times and did not present LHct levels; and (iii) anemic regular donors (IAD#2 and IAD#3), or subjects accepted for one or more donations but deferred due to LHct levels.
- After this step, we implemented a routine (P#1, in Fig. 1) that used a conversion formula for Hb to Hct to normalize the records of the attributes as follows: Hct(%) = Hb (g/dL) ∗ 3 [23–25]. In this way, we standardized and normalized the records for a single variable that did not depend on the period in which the data were collected.
- To elucidate the influence of anemia on regular blood donors, we developed a routine that inserted donation intervals (in months) and the number of blood donations into the dataset (P2#, in Fig. 1)
- The third inclusion criterion (IC#3, in Fig. 1) based on the second declaration was also implemented. In this third inclusion criterion, we selected only the dataset of anemic regular donors (IAD#4) that were not deferred due to LHct in the first recorded donation..
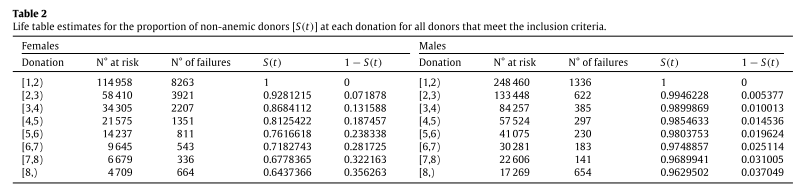
The proportion of the nonanemic population can be interpreted as a probability of the donor developing anemia up to each donation. For instance, in the fourth donation (third row of Table 2) S(3) represents the probability that a donor is non-anemic after 3 donations (but prior to the fourth donation)
Mean Hct values for the various donor groups as well as mean time intervals between donations and relative and absolute frequencies of the occurrence of anemia by gender, age and time since last donation.
Multivariate analysis results (with occurrence of anemia as the response variable):
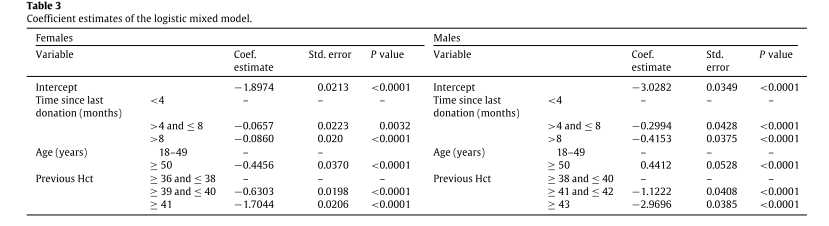
the corrected p values were 0.0008 for females and 0.5000 for males. This result shows that the random effects must be included for female donors, but they are not needed for males. Table 3 shows the coefficient estimates of the logistic final model (logit link function) for males and females. All coefficients in Table 3 are significant.
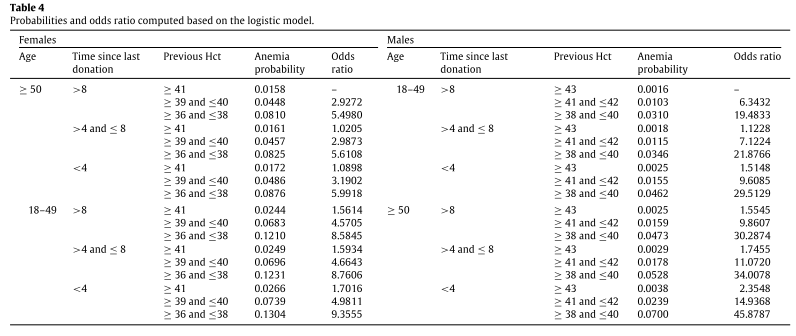
We show the values for all possible combinations of the categories of the variables (resulting in a total of 18 groups) in Table 4. For all groups, the odds ratios are computed by considering the first group as a reference. The reference group was chosen as the group with a lower anemia probability so that the odds ratio interpretation is straightforward. Note that the reference group is different for males and females. For both female and male donors, lower levels of previous Hct increases the probability of anemia in a given donation.
For both female and male donors, lower levels of previous Hct increases the probability of anemia in a given donation.
This paper shows that the provenance model is important for the generation of a reduced dataset and to provide efficient and conclusive analyses based on expert knowledge. The provenance was used to extract and select knowledge from a huge data volume.
Amanqui, Flor K.; Nies, Tom de; Dimou, Anastasia; Verborgh, Ruben; Mannens, Erik; van de Walle, Rik; Moreira, Dilvan
A Model of Provenance Applied to Biodiversity Datasets
2016
Researchers from Ghent University’s Data Science Lab and University of Sao Paulo’s ICMC contributed to this paper in the context of biodiversity research.
The Goal is a conceptual provenance model for for species identification. A primary requirement is interoperability in heterogeneous environments such as the web.
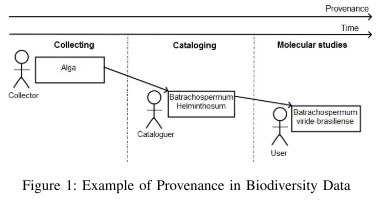
The Authors describe provenance as the “history [...] of the species”, meaning the process of species identification which typically involves different persons possibly far apart in space-time.
Methods:
- The W3C PROV ontology is used as the basis for the model https://www.w3.org/TR/prov-overview/; current version: https://www.w3.org/TR/2013/NOTE-prov-overview-20130430/)
- A mapping from ontologies, prov. model, taxonomy and collection DB to RDF (using the RML language) is defined for, and OWL is used to represent, the biodiversity data (RDF enables queries based on relationships using SPARQL). The mapping consist of “Logical Source” (dataset), “Subject Map” (resource URIs) and optional “Predicate-Object Maps”
- GeoSPARQL ontology terms and Well-Known Text (WKT) are used to describe georeferenced data
- 5 biodiversity scientists were asked to categorize key biodiversity data information like what, how and where was collected in the form of use cases (https://www.researchgate.net/publication/301287330_Interviews) -- the information resulting from the interviews was used for creating and validating the model (cf. results, Fig 3)
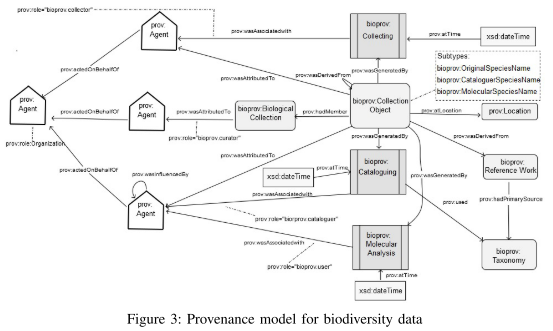
A conceptual model for provenance in biodiversity data for species identification.
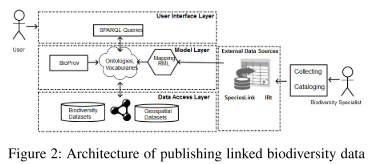
An architecture for publishing “linked biodiversity data”:
“[The] provenance model for biodiversity data, as shown in Figure 3 [ …,] defines a set of starting point terms divided into three classes: Entity, Agent and Activity. These classes are associated by relations such as prov:wasAttributedTo, prov:wasInformedBy, etc. The entity responsible for commanding the execution of an activity is modeled as an Agent.”
“In order to model the species names that are defined by the agents, [the authors] propose the following extensions [to W3C PROV] ]that are subtypes of prov:Entity:
- bioprov:OriginalSpeciesName: denotes an original species name that is not derived from any other name and the collector who emitted it is the initiator of the identification process.
- bioprov:CataloguerSpeciesName: denotes a species name which is based on another name that has been published in the past. The Cataloguer agent emitted this name based on the original species name.
- bioprov:MolecularSpeciesName: denotes a species name that is produced by modifying an existing species name. It is possible that the species name is altered. With these three extensions [the possibility that] species names could change over time [is covered]. [The] model is available at https://www.researchgate.net/publication/299690682_BioProv.”
Use cases, with one example presented in the paper.
“In this work, we presented a model for biodiversity data provenance (BioProv). BioProv enables applications that analyze biodiversity to incorporate provenance data in their information. We use the provenance information to allow experts in biodiversity to perform queries and answer scientific questions.”
No precision/recall evaluation of the queries is performed, although planned for later work.
Groth, Paul; Miles, Simon; Moreau, Luc
A Model of Process Documentation to Determine Provenance in Mash-Ups
2009
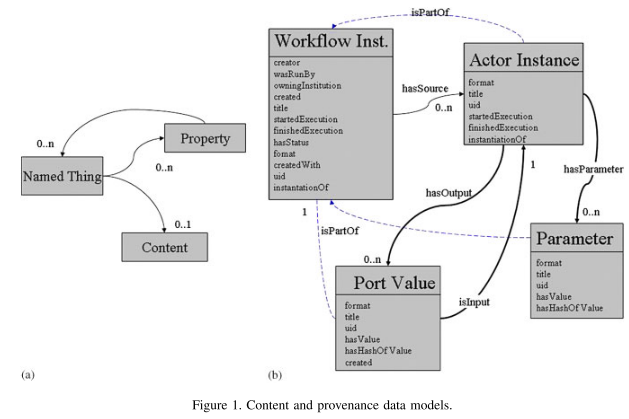
Our previous work has focused on the architecture, performance, and uses of provenance systems not on a detailed description of the data model that makes these systems possible. Furthermore, while previous work has discussed an XML instantiation of the data model, it has not described fundamental principles and design rationale behind the data model. This article describes a conceptual data model for process documentation that is independent from any one implementation or instantiation.
Process documentation is information that describes a process that has occurred. To ensure that process documentation can be created by multiple software components, across multiple institutions, in multiple domains and then used in trailing analyses, we have developed a generic data model for process documentation. [...] These properties have been evidenced by the model’s use in a variety of domains [...] This article focuses on describing this generic data model, its contributions are twofold:
(1) A precise conceptual definition of a generic data model for process
documentation. (2) An evaluation of the model with respect to a use case from the bioinformatics
domain.
“As users become reliant on these applications, they would like to understand where, why, and how their results were produced. Such questions can be answered by performing a trailing analysis that would determine the provenance of the result in question, where provenance is defined as the process that led to a result.”
- First, we motivate a generic data model for process documentation
- and enumerate a number of nonfunctional requirements that such a model should take into account.
- Factual
- Attributable
- Autonomously Creatable
- Use case: Amino acid Compressibility Experiment (ACE) as an example mash-up for evaluation of the proposed model (p-structure) -- present the various concepts that underpin the p-structure using examples from the ACE.
- We then describe a bioinformatics mash-up that foreshadows the direction of future scientific and business applications and enumerate a number of provenance questions that arise from it [DeRoure 2007]. Next, the data model and the concepts that underpin it are presented with various examples given from the use case.
- For each set of concepts, we provide a conceptmap [Novak 1998] that provides an overview ofthe concepts and the relationships between them. Concept maps were chosen for this article because they are designed for human consumption. Computer parsable representations are available as XML [Munroe et al. 2006a] and OWL (http://www.pasoa.org/schemas/ontologies/pstruct025.owl). [Relationships between concepts in the maps are read downwards or along the arrow.]
- The perspective we take is to view applications as composed ofentities, called actors, each ofwhich represents a set offunctionality within the application and interact with other actors by the sending and receiving of messages through well-defined communication endpoints.
- A qualitative evaluation of the data model with respect to the enumerated requirements and the use case’s provenance questions is then given. Finally, we discuss related work and conclude.
- Our evaluation of the data model consists of analysis of how the data model fulfils the initial requirements presented in Section 3 and a description of how the data model can be used to address the four use case questions from the ACE mash-up.
- [The actor] Collate Sample was implemented as a Java based Web Service. Compress and Calculate Efficiency were run as jobs on the Grid.
- implementations of provenance stores currently available offer XQuery [Boag et al. 2006] and a provenance specific graph traversal query function [Miles 2006]. Using these query mechanisms, the use case questions were answered.We used the PReServ provenance store in our tests [Groth et al. 2005b].
(1) A precise conceptual definition of a generic data model for process
documentation.
(2) An evaluation of the model with respect to a use case from the bioinformatics
domain.
we have defined the concepts that comprise a generic data model for process documentation, the p-structure. This description included several concept maps that unambiguously summarize the various concepts underpinning the data model and the relationships between them. The data
model supports the autonomous creation of factual, attributable process documentation
by separate, distributed application components.
P-assertions contain the elements necessary to represent a process.
We place a restriction on all asserters [actors] that they only create p-assertions for events and data that are inside their scope [req. of factuality]. [p-assertion also has an asserter identity, satisfying the req. of attributability
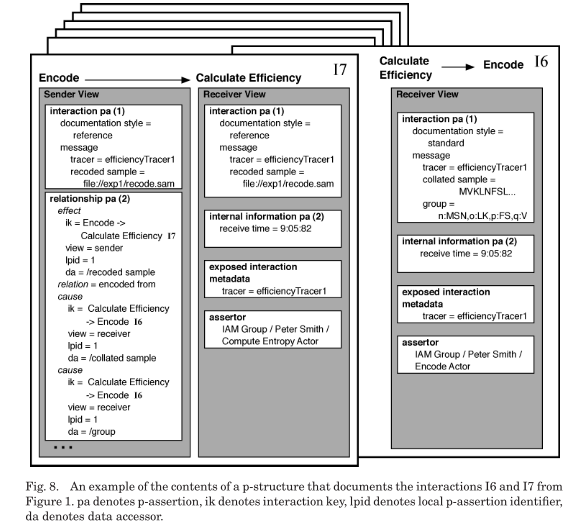
Interactions are represented by interaction p-assertions, which contain four parts:
(1) An asserter identity.
(2) An event identifier.
(3) A representation of the message exchanged in the interaction. (4) A documentation style describing how the representation was generated.
A relationship p-assertion identifies one or more occurrences [= event or data item of event] that are causes and one occurrence that is the effect of those causes. We limit a relationship p-assertion to one effect to make it easier to find the provenance of a particular occurrence
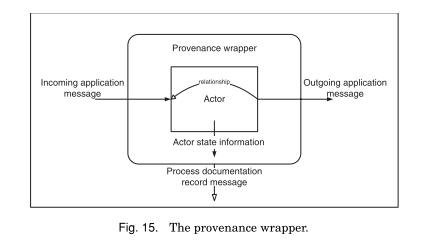
[Structural, transformational] Relationship p-assertions document both the data flow and control flow within an actor and thus are critical to understanding the process within an application.
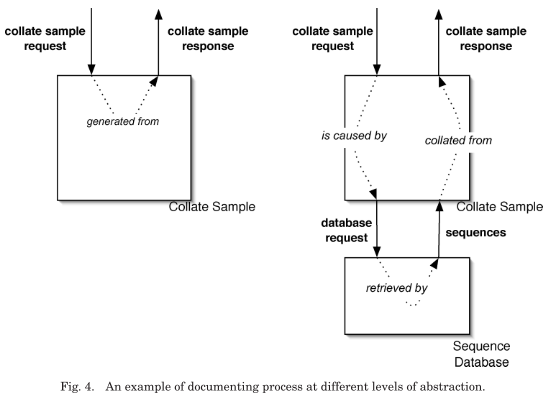
The combination of interaction p-assertions and relationship p-assertions provide the information necessary to document processes. Furthermore, they allow process documentation to be created at different levels of abstraction:
an internal information p-assertion consists of three parts: the data, the documentation style of the data, and the event identifier of the event to which the data is causally connected. Internal information p-assertions allow data items to bemade explicit without, the sometimes unnecessary, overhead of creating documentation for their generation
the p-structure [...] is an organization of p-assertions that provides several elements to help isolate, find, and understand sets of p-assertions.
p-asssertions are [placed in a view, with a local id, and] grouped together by the event that they are most closely associated with [effect event for rel. p-assert.]. [Each view is associated with an asserter]
For the independent creation req. to be satisfiable the following rules apply
- Unique [Sender] Interaction Key Rule.
- Interaction Key Transmission Rule.
- Appropriate Interaction Rule.
a graph of causal connections leading back to a particular occurrence can be extracted from the p-structure. Such a causality graph describes the provenance of an occurrence.
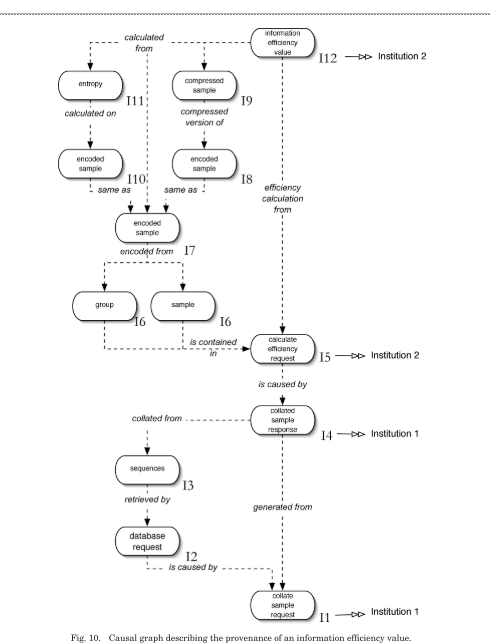
Figure 10 shows the provenance of an information efficiency value. The nodes in the graph are occurrences, in the role of causes, effects or both. The edges in the graph are hyper-edges and represent the causal connections extracted from relationship p-assertions. All arrows on the edges point from effect to cause.
(1) What were the original sequences used in generating an information efficiency value? To find the answer to this question, the provenance of the particular information efficiency value needs to be known. The following is a simplified algorithm for finding the provenance of a given occurrence, in this case it would be a particular information efficiency value. (1) All the relationship p-assertions where a given occurrence, X, was an effect are found. (2) From these relationship p-assertions, all the occurrences that were the causes of the effect can be found. (3) For each of the occurrences found in Step 2, find the provenance of those items using this algorithm. This algorithm runs until there are no more relationship p-assertions and the entire causality graph for the information efficiency value is generated as shown in Figure 10.
(2) Which institutions were involved in the production of a particular information efficiency value? … The p-structure allows p-assertions produced by multiple institutions to be collated together. Once process documentation has been assembled, this question can be answered in the same manner as the previous question, however, in this instance, the data held within internal information p-assertions is used.
(3) What were the common steps in the production of these two information efficiency values? To answer this question, a notion of a process that can be differentiated from other processes is required. [...]
Knowing the provenance of the results of multiple experimental runs and being able to differentiate and compare them is fundamental to answering this question. Therefore, the solution to this question involves finding the provenance of each information efficiency value, which generates two causal graphs. The common interactions between these two causal graphs are then found by comparing the interaction keys of the interactions. These interactions are the common steps asked for in the use case. Using tracers, we can then determine if the interactions belong to larger processes.
(4) Were references or pointers used when documenting this experiment run?
... Our data model for process documentation takes this concern into account by using documentation styles to label p-assertions that contain references.
Therefore, the solution to this use case relies on documentation styles. First, the provenance of a particular experiment run is found, then for every interaction p-assertion contained in the returned causality graph, the documentation style is retrieved. If the style retrieved is a reference documentation style, an identifier for the corresponding interaction p-assertion can be returned demonstrating that references were used in the run.
We have defined the concepts that comprise a generic data model for process documentation, the p-structure. This description included several concept maps that unambiguously summarize the various concepts underpinning the data model and the relationships between them. The data
model supports the autonomous creation of factual, attributable process documentation by separate, distributed application components.
Ma, Taotao; Wang, Hua; Yong, Jianming; Zhao, Yueai
Causal dependencies of provenance data in healthcare environment
2015
Although a general provenance model has been proposed by Ni et al [1], its main focus is on access control for provenance, and this model is not able to distinguish between the provenances in applications and system level. [...] the model proposed by Salmin et al [2] [...] does not capture casual dependencies where access control polices and decisions are based on.
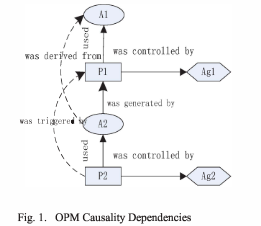
A provenance model is able to describe the provenance of any data at an abstract layer, but does not explicitly capture causal dependencies that are a vital challenge since the lacks of the relations in OPM, especially in healthcare environment. In this paper, we analyse the causal dependencies between entities in a medical workflow system with OPM graphs.
“Provenance, or history of data, or information about the origin, derivation, describing how an object came to be in its present, is becoming a more and more important topic.”
In this paper, in order to address the shortcoming in the proposed model [2], we utilize the standard model OPM [3], fix the causal dependencies of provenance data in a real workflow health care system. [...] Section 2 introduces the provenance data dependency. Section 3 introduces the related work including previously proposed models. We analyse use case models, and capture the causal dependencies in OPM graph in Section 4. Section 5 draws conclusions for work.
We also analyze the requirements that the model should be satisfied.
[An OPM model which explicitly captures causal dependencies;
a use case.]
In this paper, we first introduce the OPM model, which can capture the causal dependencies between the entities, then based on a proposed access control model for provenance capture the provenance information.
Full results omitted; see full text.
We apply the OPM model to access control the provenance information. So it can explicitly capture the causal dependencies where the access control policies and other preference based on
Schreiber, Andreas
A Provenance Model for Quantified Self Data
2016
[Unspecified.]
By capturing and storing provenance self-trackers could answer questions
regarding the creation process of his data, security aspects, and even determining
the trustfulness of his diagnostic results (Sect. 3.1). Like any database management
system, a provenance system needs an underlying data model (provenance
data model) that defines the data that will be captured in QS workflows.
“To answer those questions and to understand how the data of the user is created, manipulated and processed, the provenance [15] of that data can be recorded and analyzed. Provenance has many synonyms like lineage, genealogy or pedigree that results from different domains in which they were elaborated separately until a few years ago.”
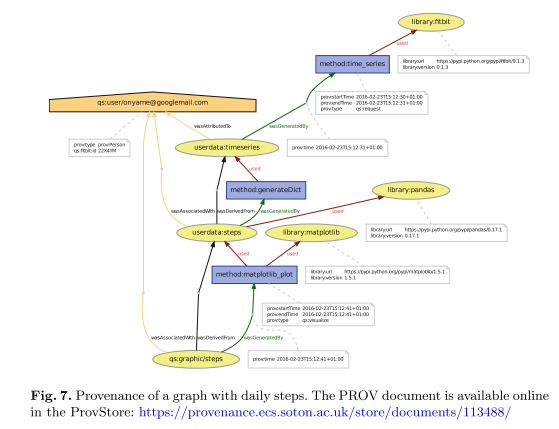
Both are based on the open W3C standards PROV-O and PROV-DM. We show the feasibility of the presented provenance model with a small workflow using steps data from Fitbit fitness tracker.
[Unspecified; but formal methods of description-logic-based modelling are used (Protege, OWL).]
[A model (descriptive and partly formalised);
use case based evaluation (visualisation Fitbit): informal assessment.]
We present a solution that helps to get insight in Quantified Self data flows and to answer questions related to data security. We provided an ontology and a provenance data model for Quantified Self
data.
[The model works according to qualitative evaluation.]
Full results omitted; see full text.
Using that model, developers and users can record provenance of Quantified Self apps and services with a standardized notation. We show the feasibility of the presented provenance model with a small workflow using steps data from Fitbit fitness tracker.
Zhao, J.; Wroe, C.; Goble, C.; Stevens, R.; Quan, D.; Greenwood, M.
Using semantic web technologies for representing e-Science provenance
2004
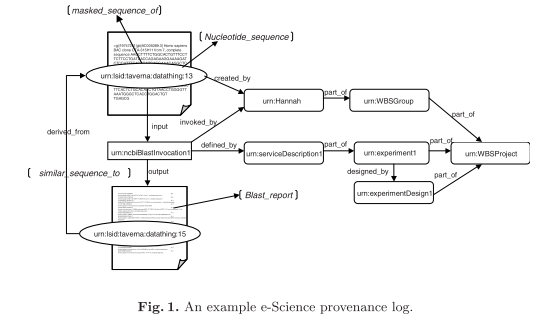
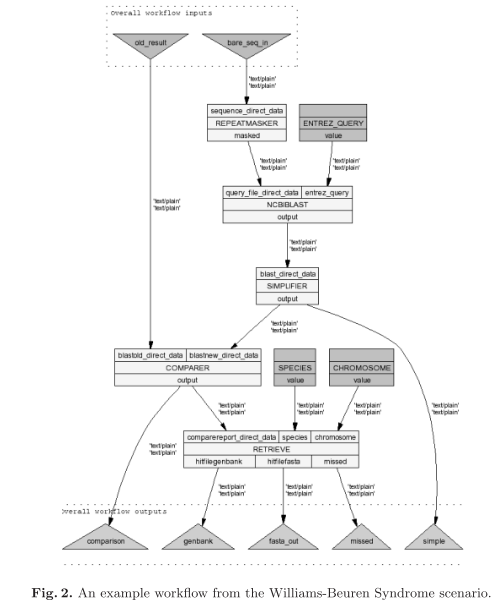
myGrid 1, as a pilot e-Science project in the U.K., aims to provide middleware services not only to automate the execution of in silico experiments as workflows in a Grid environment, but also to manage and use results from experiments [4]. Currently this project is being developed using several molecular biological scenarios.
This paper uses the collaboration with biologists and bioinformaticians at Manchester University, researching the genetic basis of Williams-Beuren Syndrome [5].
This work therefore forms a case study showing how existing Semantic Web tools can effectively support the emerging requirements of life science research.
“Traditionally, in the lab, a scientist records such information in a lab-book and uses these records of where, how and why results were generated in the process of analysing, validating and publishing scientific findings. These records are the provenance of an experiment.”
“Provenance has a very broad scope within e-Science. At its simplest, it describes from where something has arisen.”
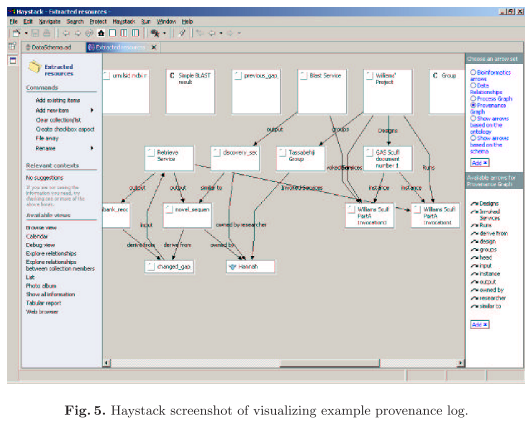
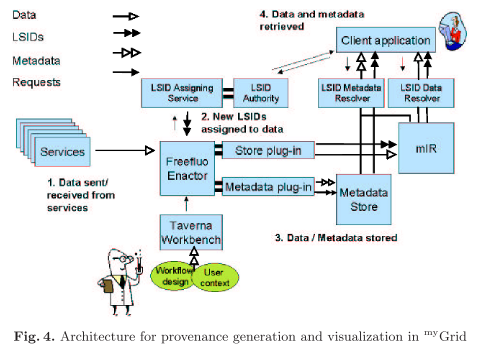
By using existing Semantic Web technology and software such as RDF repositories, ontologies, Haystack and Life Science
Identifiers, this work acts as a case study describing what is already possible in supporting life scientists with a Semantic Web. In this paper Sect 2 describes scientists’ requirements for provenance and the implications of the information recorded. [...]
Therefore we have explored the use of Semantic Web technologies such as RDF, and ontologies to support its representation and used existing initiatives such as Jena and LSID, to generate and store such material. Haystack has been used to provide multiple views of provenance metadata that can be further annotated.
This work therefore forms a case study showing how existing Semantic Web tools can effectively support the emerging requirements of life science research.
This work therefore forms a case study showing how existing Semantic Web tools can effectively support the emerging requirements of life science research.
Full results omitted; see full text.
Given our addition of a semantic description of these data, we fully expect to
be able to add machine processing of these data to the current human emphasis
of our work. [...] This would allow
semantic querying of these logs, provide trust information about data based on
logs, etc.
Zhong, Han; Chen, Jianhui; Kotake, Taihei; Han, Jian; Zhong, Ning; Huang, Zhisheng
Developing a Brain Informatics Provenance Model
2013
[Unspecified.]
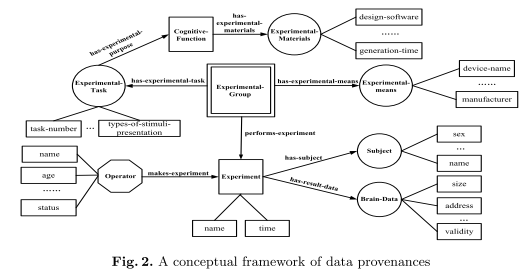
[...] Such a provenance model facilitates more accurate modeling of brain data, including data creation and data processing for integrating various primitive brain data, brain data related information during the systematic Brain Informatics study.
“Provenance information describes the origins and the history of data in its life cycle and has been studied based on the relational database, XML,etc [1,3,8].”
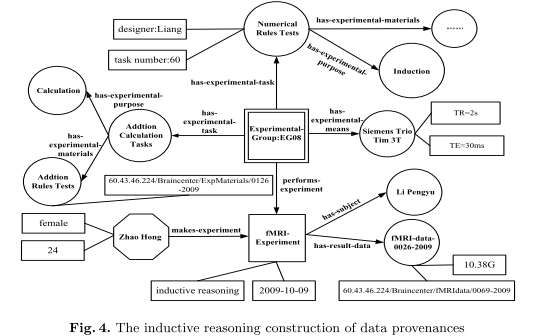
In this paper we put forward a provenance model for constructing BI provenances. Sections 3 and 4 describe such a provenance model and its conceptual framework, respectively. Section 5 provides a case study in thinking-centric systematic investigation. [...]
model elements are identified and defined by extending the Open Provenance Model. A case study is also described to
demonstrate significance and usefulness of the proposed model.
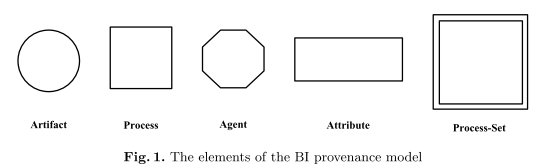
[A] provenance model
of brain data […] A case study is also described to demonstrate significance and usefulness of the proposed model[.]
This paper puts forward a provenance model of brain data[...]
Full results omitted; see full text.
Such a provenance model facilitates more accurate modeling of brain data, including data creation and data processing for integrating various primitive brain data, brain data related information during the systematic Brain Informatics study.
Balis, Bartosz; Bubak, Marian; Wach, Jakub
Provenance tracking in the ViroLab virtual laboratory
2008
The goal of ViroLab1 [11] is to set up a virtual organization and develop a virtual laboratory2 for infectious diseases in order to support medical doctors and scientists in their daily work. The primary scenario that drives ViroLab is the HIV drug resistance. Medical doctors are provided with a decision support system which helps them rank drugs for particular patients. Virologists can use data integrated from a number of institutions, and various tools, virtualized and perhaps composed into scripts (similar to workflows), to perform data mining, statistical analysis, as well as molecular dynamics and cellular automata simulations.
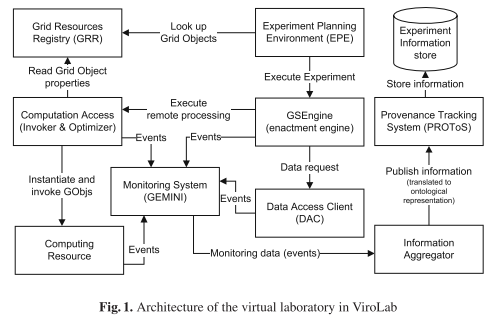
This paper presents the provenance tracking approach in the ViroLab Project.
R. Wyrzykowski et al. (Eds.): PPAM 2007, LNCS 4967, pp. 381–390, 2008.
Keywords: e-Science, Grid, ontology, provenance, ViroLab.
Bartosz Bali´s1,2, Marian Bubak1,2, and Jakub Wach2 1 Institute of Computer Science, AGH, Poland {balis,bubak}@agh.edu.pl, wach.kuba@gmail.com 2 Academic Computer Centre – CYFRONET, Poland
The goal of ViroLab1 [11] is to set up a virtual organization and develop a virtual laboratory2 for infectious diseases in order to support medical doctors and scientists in
their daily work. The primary scenario that drives ViroLab is the HIV drug resistance. Medical doctors are provided with a decision support system which helps them rank drugs for particular patients. Virologists can use data integrated from a number of institutions, and various tools, virtualized and perhaps composed into scripts (similar to workflows), to perform data mining, statistical analysis, as well as molecular dynamics and cellular automata simulations.
This paper presents the provenance tracking approach in the ViroLab Project.
Our motivation in designing the provenance solution was not only to support recording and browsing of provenance logs but also to enable complex queries over provenance data that can help find interesting properties. Examples of useful queries are as follows: – Which rule sets were used most frequently to obtain drug rankings? – Return all experiments performed by John Doe within last two weeks. – Return all input sequences of experiments of type Drug Ranking Support whose input parameter threshold was between 0.03 and 0.04.
Our work is also driven by the vision of semantic grid as a future infrastructure for e-Science, as presented in [10]. In this article, three types of services – data services, information services, and knowledge services – are introduced, where data is understood as an uninterpreted sequence of bits (e.g. an integer value), information is data associated with meaning (e.g. a temperature), while knowledge is understood as “information applied to achieve a goal, solve a problem or enact a decision”. Provenance is important part of an e-Science infrastructure providing both information and knowledge services. However, an adequate knowledge management has several aspects concerning various stages of knowledge lifecycle: acquisition, modelling, retrieval, reuse, publishing and maintenance [10]. Consequently, our aim is to design provenance data model and representation as well as the provenance tracking system with those requirements in mind.
With the provenance service built as a knowledge service, its use can go beyond the simple usage as a personal scientist’s logbook documenting the experiments. Sufficiently rich information stored in provenance records can enable other uses. For example, records of multipe invocations of particular services equipped with timing information can easily be used for instance-based learning to optimize future resource brokering or scheduling decisions. In the future, we anticipate the integration with the ViroLab’s brokering/scheduling component for this purpose.
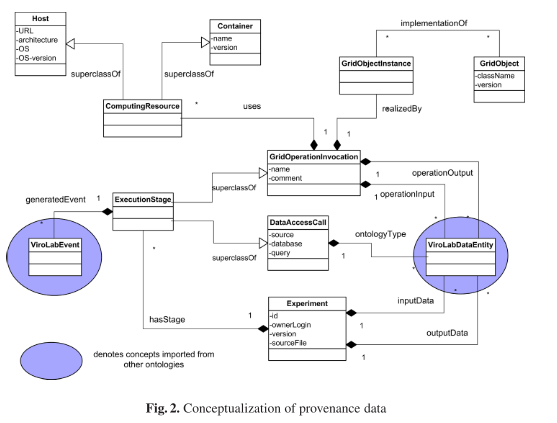
“Provenance of a piece of data is defined as a process that led to that data [5], also known as a derivation path of a piece of data.”
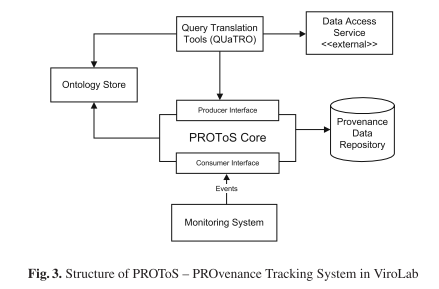
This paper presents the provenance tracking approach in the ViroLab Project. We describe our motivation (Section 2), the provenance model in ViroLab (Section 3), and the architecture of our provenance tracking system – PROToS (Section 4). Section 5 presents the implementation of PROToS in more detail. Section 6 contains an overview of related work.
The applied provenance solution is motivated by the Semantic Grid vision as an infrastructure for e-Science. Provenance data is represented in XML and modeled as ontologies described in the OWL knowledge representation language.
[Unspecified.]
[A] provenance tracking system[.]
We present a provenance tracking approach developed as part of the virtual laboratory of the ViroLab project.
Full results omitted; see full text.
Butin, Denis; Demirel, Denise; Buchmann, Johannes
Formal Policy-Based Provenance Audit
2016
[Unspecified.]
K. Ogawa and K. Yoshioka (Eds.): IWSEC 2016, LNCS 9836, pp. 234–253, 2016.
Denis Butin(B), Denise Demirel, and Johannes Buchmann
TU Darmstadt, Darmstadt, Germany {dbutin,ddemirel,buchmann}@cdc.informatik.tu-darmstadt.de
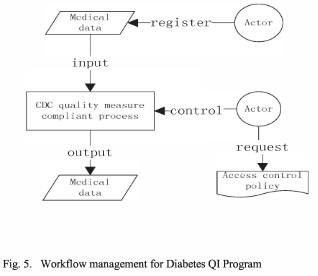
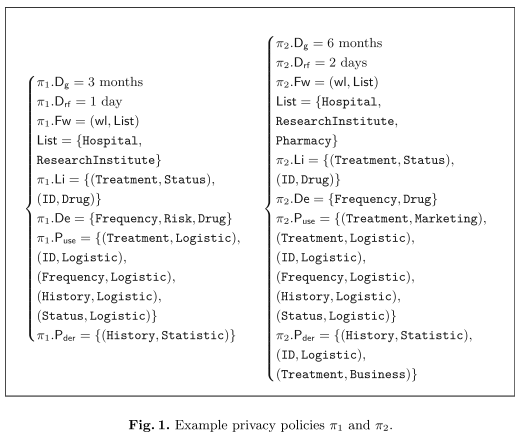
While the idea of using provenance as a tool to audit the compliance with privacy regulations is not new, so far formal approaches have been lacking. Both the consistency of provenance records and their compliance with associated privacy policies ought to be stated precisely to pave the way for automated analyses. Some aspects such as the enforcement of processing purposes are not fully amenable to automation, but provenance records allow to collect enough information for complementary manual verification. Contribution. We present a formal framework modelling provenance events and their compliance with respect to usage policies.
“In this work, we adopt the W3C perspective on provenance as (...) a record that describes the people, institutions, entities and activities, involved in producing, influencing, or delivering a piece of data or a thing [24]. Hence the key idea of provenance—as taken up here—is to paint a comprehensive picture clarifying the life cycle of data, from its creation to its destruction, including the way it is used, and how other data is derived or linked from it.”
Contribution. We present a formal framework modelling provenance events and their compliance with respect to usage policies. We first introduce usage policies and model them as tuples including forwarding policies, authorised purposes for different types of operations, deletion delays, and linking and derivation policies (Sect. 2.1). A data category is not necessarily always associated with the same usage policy. The relevant policy depends on the component and on the log events under consideration. Afterwards, we model provenance records as sequences of discrete events. Each record refers to a single data subject, e.g. a patient in the eHealth scenario, but can accommodate an arbitrary number of entities and processed data categories. The granularity of the events is chosen in accordance with the policies (Sect. 2.2). Next, we establish additional notation (Sect. 3.1) and formalise the correctness (Sect. 3.2) and compliance of logs of provenance events when dealing with privacy policies (Sect. 3.3). Correctness pertains to the internal consistency of the provenance record, independently of the usage policies under consideration. Conversely, compliance relates to the relation between the provenance record and the associated usage policies. This formalisation can serve as a basis for a posteriori provenance analysis by an auditor. Next, we discuss the limitations of the approach when dealing with privacy policies and show how it can be included in a global accountability process (Sect. 4). We consider the case of privacy separately because the guarantees at stake are especially critical when personal data is involved. Furthermore, the application of provenance records to this use case immediately raises the question of whether logging creates additional privacy risks. We show under which conditions our framework for provenance-based auditing is applicable to privacy policies. The framework is then evaluated through a scenario involving personal medical data processed among different entities (Sect. 5). Finally, we provide a review of related work (Sect. 6) and offer our conclusions (Sect. 7).
The practical applicability of our approach is demonstrated using a provenance record involving medical data and corresponding privacy policies with personal data protection as a goal. We have introduced a formal correctness and compliance framework for provenance,
based on a linear view of provenance events and the definition of sticky
usage policies.
[A formal framework for
policy-based provenance audit;
use case based evaluation (personal data protection): informal assessment.]
In this paper, we introduce a formal framework for policy-based provenance audit. We show how it can be used to demonstrate correctness, consistency, and compliance of provenance records with machine-readable usage policies. We also analyse the suitability of our framework for the special case of privacy protection.
Full results omitted; see full text.
The presented format for policies and rules for correctness and compliance are not meant to be exhaustive or applicable to all scenarios. However, our framework serves as a basis allowing to include other policy components and event types. Therefore, the choices presented here in terms of policy components,
event types, and rules should only be seen as an instantiation of the framework. A scenario involving the processing of medical data by different, communicating data controllers was used to exemplify the framework. The use of such a compliance framework is only meaningful within a wider accountability process, since provenance records cannot be intrinsically trustworthy. In combination with such an accountability process it can however contribute to increase transparency about personal data handling, ultimately benefiting data subjects.
Groth, P.; Luck, M.; Moreau, L.
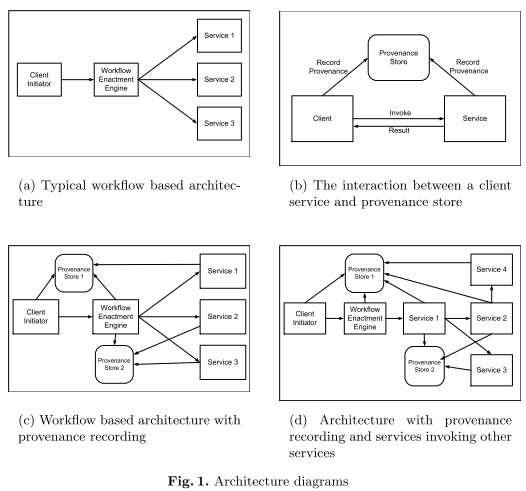
A protocol for recording provenance in service-oriented grids
2005
In summary, the paucity of standards, components, and techniques for recording provenance is a problem that needs to be addressed by the Grid community. The focus of this work, the development of a general architecture and protocol for recording provenance, is a first step towards addressing these problems.
T. Higashino (Ed.): OPODIS 2004, LNCS 3544, pp. 124–139, 2005.
Keywords: recording provenance, provenance, grids, web services, lineage.
Paul Groth, Michael Luck, and Luc Moreau School of Electronics and Computer Science, University of Southampton, Highfield, Southampton SO17 1BJ, United Kingdom {pg03r, mml, l.moreau}@ecs.soton.ac.uk
Given the need for provenance information and the emergence of Grids as infrastructure for running major applications, a problem arises that has yet to be fully addressed by the Grid community, namely, how to record provenance in Grids? Some bespoke and ad-hoc solutions have been developed to address the lack of provenance recording capability within the context of specific Grid applications. Unfortunately, this means that such provenance systems cannot interoperate. Therefore, incompatibility of components prevents provenance from being shared. Furthermore, the absence of components for recording provenance makes the development of applications requiring provenance recording more complicated and onerous.
Another drawback to current bespoke solutions is the inability for provenance to be shared by different parties. Even with the availability of provenancerelated software components, the goal of sharing provenance information will not be achieved. To address this problem, standards should be developed for how provenance information is recorded, represented, and accessed. Such standards would allow provenance to be shared across applications, provenance components, and Grids, making provenance information more accessible and valuable. In summary, the paucity of standards, components, and techniques for recording provenance is a problem that needs to be addressed by the Grid community. The focus of this work, the development of a general architecture and protocol for recording provenance, is a first step towards addressing these problems.
“We define the provenance of some data as the documentation of the process that led to the data.”
The rest of the paper is organised as follows: Section 2 presents a set of requirements that a provenance system should address. Then, Section 3 outlines a design for a provenance recording system in the context of service-oriented architectures. The key element of our system is the Provenance Recording Protocol described in Section 4. In Section 5, the actors in the system are formalised, and the formalisations are then used, in Section 6, to derive some important properties of the system. Finally, Section 7 discusses related work, followed by a conclusion. Given the length of this paper, we assume the reader is familiar with Grids, Virtual Organisations (VO), Web Services, and service-oriented architectures (SOA).
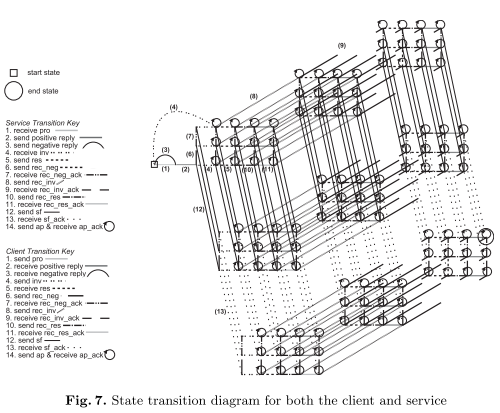
We describe the protocol in the context of a service-oriented architecture and formalise the entities involved using an abstract state machine or a three-dimensional state transition diagram. Using these techniques we sketch a liveness property for the system.
[A]n implementation-independent protocol for
the recording of provenance.
To support provenance capture in Grids, we have developed an implementation-independent protocol for the recording of provenance.
Full results omitted; see full text.
The necessity for storing, maintaining and tracking provenance is evident in fields ranging from biology to aerospace. As science and business embrace Grids as a mechanism to achieve their goals, recording provenance will become an ever more important factor in the construction of Grids. The development of common components, protocols, and standards will make this construction process faster, easier, and more interoperable. In this paper, we presented a stepping stone to the development of a common provenance recording system, namely, an implementation-independent protocol for recording provenance, PReP
Portokallidis, Nick; Drosatos, George; Kaldoudi, Eleni
Capturing Provenance, Evolution and Modification of Clinical Protocols via a Heterogeneous, Semantic Social Network
2016
[Unspecified.]
Nursing Informatics 2016 W. Sermeus et al. (Eds.)
Keywords. Clinical protocols, semantic social network, ontology, provenance.
Nick PORTOKALLIDIS, George DROSATOS and Eleni KALDOUDI1
School of Medicine, Democritus University of Thrace, Alexandroupoli, Greece
Research so far has rigorously addressed the computerized execution of clinical protocols and this has resulted in a number of related representation languages, execution engines and integrated platforms to support the real time execution [4], [5]. However, much less effort has been put into organizing available clinical protocols. Mainly, they are maintained in data silos of the respective issuing body without means for straightforward seamless integration and open availability.
In this work we build on the paradigm of social associations among human and non-human entities alike and propose a novel approach to describe and organize clinical protocols for easy use and reuse. The following sections discuss different perspectives of clinical protocol provenance, evolution and modification and present a novel approach for organizing, and managing clinical protocols via a heterogeneous, semantic social network.
“The clinical protocol origin is of outmost importance for a number of reasons. The first is provenance: no one could (or should) trust data purporting to represent medical knowledge without the ability to trace it back to its source.”
In this work we build on the paradigm of social associations among human and non-human entities alike and propose a novel approach to describe and organize clinical protocols for easy use and reuse. The following sections discuss different perspectives of clinical protocol provenance, evolution and modification and present a novel approach for organizing, and managing clinical protocols via a heterogeneous, semantic social network.
[Unspecified.]
[A] heterogeneous semantic social network to describe and organize clinical protocols based on their provenance, evolution and modifications.
In this paper we propose a heterogeneous semantic social network to describe and organize clinical protocols based on their provenance, evolution and modifications. The proposed approach allows semantic tagging and enrichment of clinical protocols so that they can be used and re-used across platforms and also be linked directly to other relevant scientific information, e.g. published works in PubMed or personal health records, and other clinical information systems.
Full results omitted; see full text.
Schuchardt, Karen L.; Gibson, Tara; Stepban, Eric; Chin Jr., George
Applying content management to automated provenance capture
2008
In our work developing problem solving and collaboration environments, we make extensive use of provenance and metadata to annotate data objects and capture their relationship to other objects.
Prior to the issuing of the Provenance Challenge [3], we were engaged in a prototype research effort to combine several components: workflow systems, scientific content management, advanced query languages, and a provenance data model to define a general provenance architecture in the context of automated workflow. To demonstrate the applicability and viability of workflow/provenance systems, we collaborated with biologists at PNNL to simulate analyzing and visualizing public protein–protein interaction databases against Microarray data sets. This effort led to multiple extensions of our architecture including enhanced indexing, translation services, and naming services. With our preliminary results in place, the Provenance Challenge provided an ideal environment to further validate our approach and compare it to other leading approaches. To put our solution within the context of the overall Challenge and guide the reader interested in comparing approaches, Table I provides a listing of the sections pertaining to our solution and a cross reference to other provenance participants that use similar approaches.
CONCURRENCY AND COMPUTATION: PRACTICE AND EXPERIENCE Concurrency Computat.: Pract. Exper. 2008; 20:541–554
KEY WORDS: provenance; metadata; workflow; content management
Karen L. Schuchardt∗, †, Tara Gibson, Eric Stephan and George Chin Jr
Pacific Northwest National Laboratory, P.O. Box 999, K7-90, Richland, WA 99352, U.S.A.
Like other recent provenance research efforts [19–21], our research focuses on developing mechanisms for representing, collecting, storing, and disseminating provenance information on scientific data products. Our goal is to be able to work with any workflow execution environment. To store provenance, metadata, and data artifacts, we apply and extend our existing content management systems which naturally supports how scientists work and manage their data.
“Data provenance—the metadata that captures the ‘derivation history of a data product starting from its original sources [2]’—and the tools and technologies that support it play a crucial role in enabling scientists to effectively access and use ever-growing scientific data and to unlock and discover the knowledge encompassed within.”
We applied this prototype to the provenance challenge to demonstrate an end-to-end system that supports dynamic provenance capture, persistent content management, and dynamic searches of both provenance and metadata. We
describe our prototype, which extends the Kepler system for the execution environment, the Scientific
Annotation Middleware (SAM) content management software for data services, and an existing HTTPbased
query protocol.
[Description of a workflow provenance system prototype, Scientific Annotation Middleware (SAM) content management software for data services, and an existing HTTPbased query protocol.]
We have been prototyping a workflow provenance system, targeted at biological workflows, that extends our content management technologies and other open source tools. Our implementation offers several unique capabilities, and through the use of
standards, is able to provide access to the provenance record with a variety of commonly available client
tools.
Full results omitted; see full text.
With these extensions we have a complete, end-to-end provenance capture, storage, and query solution that features: a configurable provenance capture plug-in where individual processes as well as the workflow engine, can contribute provenance and metadata; a persistent, web protocol-based content storage solution that supports automatic metadata extraction for registered data types; an RDF-based provenance data model; query optimization by indexing both content and metadata using the Lucene index system; a bi-directional index on all provenance for forward, reverse, or mixed (both forward and reverse) direction searches, and a web protocol-based query language (DASL/SEDASL) with an extended grammar to enable users to specify SPARQL-like query criteria. In comparison with the results to other Provenance Challenge participants (Table I), we found that like most projects, we chose a workflow execution environment and like several other projects, we used the Kepler workflow technology which in many ways is comparable to Taverna in that the workflow engine provides a graphical user interface (GUI) for designing and operating workflow. Although we used a distinct representation technology, we did make use of RDF as a mechanism to model provenance, which appears to be a pretty common approach. Our query language, SEDASL seemed to hold many similarities to SPARQL and RDQL. The SEDASL query extensions provide a flexible mechanism to traverse arbitrary semantically linked provenance information. The capability to specify the output format and the support of GXL makes certain types of analysis easier to implement. However, because the query language is in XML, it is quite verbose, making it, at times difficult to read, when compared to SPARQL (http://www.w3.org/TR/rdf-sparql-query/) or other RDF query languages. As many of the ideas behind SEDASL and SPARQL are similar, we have looked at the possibility of extending our query interface to also support SPARQL (or another similar language). Much of the language, such as the metadata properties and literals could be mapped to the underlying SAM query implementation. The biggest difference between SEDASL and SPARQL is the idea of ‘variables’, however, they both represent a similar, graph-like structure. Because of this, the mapping should be possible. However, our intention is to investigate merging an RDF store with enhanced SPARQL or iTQL support with our content management and data services. Copyright q 2007 John Wiley & Sons, Ltd. Concurrency C
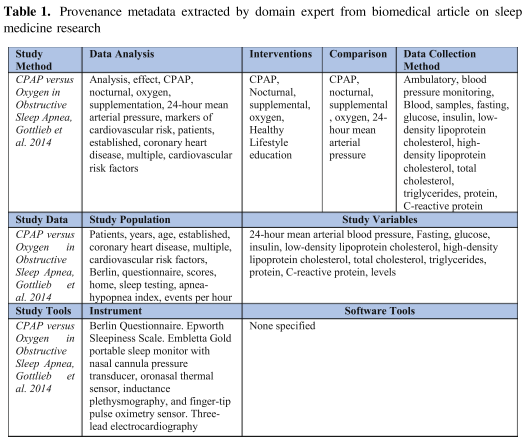
Valdez, Joshua; Rueschman, Michael; Kim, Matthew; Redline, Susan; Sahoo, Satya S.
An Ontology-Enabled Natural Language Processing Pipeline for Provenance Metadata Extraction from Biomedical Text
2016
As part of a NIH funded Big Data to Knowledge (BD2 K) project on data provenance, we have defined the Provenance for Clinical and Healthcare Research (ProvCaRe) framework that extracts, analyzes, and stores provenance of research studies to support scientific reproducibility.
The ProvCaRe framework is being developed in close collaboration with biomedical domain experts involved with the National Sleep Research Resource (NSRR), which is the largest data repository of publicly available research sleep medicine studies in the world, with a goal of aggregating more than 50,000 studies collected from 36,000 participants [4]. Using this dataset will ensure that the ProvCaRe framework reflects the practical requirements of biomedical domain and it is also scalable with increasing volume and variety of biomedical data. The NSRR project has publicly released data from more than six sleep research studies and we use peer-reviewed publications associated with these studies to extract, model, and analyze provenance information associated with these research studies.
C. Debruyne et al. (Eds.): OTM 2016 Conferences, LNCS 10033, pp. 699–708, 2016.
Keywords: Ontology-based natural language processing
metadata Scientific reproducibility Named entity recognition
Provenance
Joshua Valdez1, Michael Rueschman2, Matthew Kim2, Susan Redline2, and Satya S. Sahoo1(&) 1 Division of Medical Informatics and Electrical Engineering and Computer Science Department, Case Western Reserve University, Cleveland, OH, USA {joshua.valdez,satya.sahoo}@case.edu
2
Departments of Medicine, Brigham and Women’s Hospital and Beth Israel Deaconess Medical Center, Harvard University, Boston, MA, USA {mrueschman,mikim,sredline1}@bwh.harvard.edu
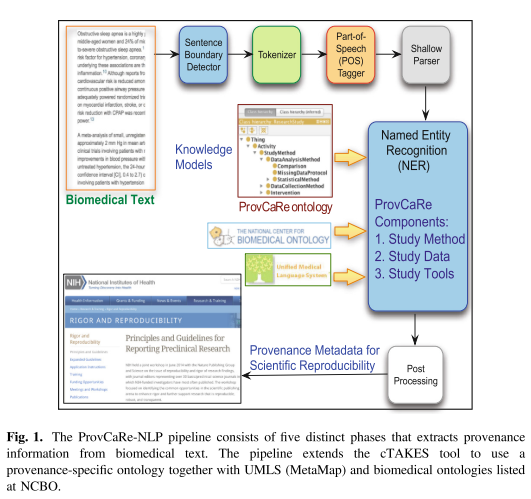
Provenance metadata describing the contextual information of scientific experiments can facilitate the evaluation of the soundness and rigor of research studies [1]. In addition, provenance metadata can support scientific reproducibility, which is one of the core foundations of scientific advancement [2]. The importance of scientific rigor and reproducibility is highlighted by the recent concerted efforts led the US National Institutes of Health (NIH) called “Rigor and Reproducibility” to ensure that results from research studies can be reproduced by other researchers in the community [2, 3]. As part of a NIH funded Big Data to Knowledge (BD2 K) project on data provenance, we have defined the Provenance for Clinical and Healthcare Research (ProvCaRe) framework that extracts, analyzes, and stores provenance of research studies to support scientific reproducibility.
The ProvCaRe framework is being developed in close collaboration with biomedical domain experts involved with the National Sleep Research Resource (NSRR), which is the largest data repository of publicly available research sleep medicine studies in the world, with a goal of aggregating more than 50,000 studies collected from 36,000 participants [4]. Using this dataset will ensure that the ProvCaRe framework reflects the practical requirements of biomedical domain and it is also scalable with increasing volume and variety of biomedical data.
The key challenges in extracting structured data from biomedical text are the use of domain-specific terminology, complex sentence structure including use of negation and modifiers that significantly modify the meaning of terms or phrases in a sentence, and identification of complex relations that link terms across one or more sentences [5].
“Provenance metadata describing the contextual information of scientific experiments”
The rest ofthe paper is organized as follows. Section 2 reviews related work. Section 3 discusses the provenance model layer of RDFPROV and introduces a sample provenance ontology. Sections 4, 5, and 6 present the model mapping layer of RDFPROV, elaborating on provenance ontology to database schema mapping, provenance metadata to relational data mapping, and SPARQLto-SQL query translation, respectively. Section 7 provides our case study for provenance management in a real-life scientific workflow from the biological simulations field. Section 8 empirically compares RDFPROV with two commercial relational RDF stores. Finally, Section 9 concludes the paper and discusses possible future work directions.
[Unspecified.]
[An] approach to extract provenance metadata from published biomedical research literature,
[evaluated] using a corpus of 20 peer-reviewed publications.
Full results omitted; see full text.
Biton, Olivier; Cohen-Boulakia, Sarah; Davidson, Susan B.; Hara, Carmern S.
Querying and managing provenance through user views in scientific workflows
2008
[Unspecified.]
ICDE 2008
Olivier Biton #1, Sarah Cohen-Boulakia #**2 Susan B. Davidson #3, Carmem S. Hara *4 # University of Pennsylvania, Philadelphia, USA {lbiton, 2sarahcb, 3susan}@cis.upenn.edu ** Universite' Paris-Sud 11, Orsay, France * Universidade Federal do Parana, Brazil 4carmem@inf.ufpr.br
In order to understand and reproduce the results of an experiment, scientists must be able to determine what sequence of steps and input data were used to produce data objects, i.e. ask provenance queries, such as: What are all the data objects! sequence of steps which have been used to produce this tree?
However, since a workflow execution may contain many steps and data objects, the amount of provenance information can be overwhelming.
There is therefore a need for abstraction mechanisms to present the most relevant provenance information to the user.
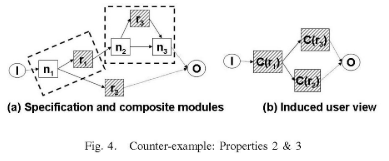
A technique that is used in systems such as Kepler [1] and Taverna [3] is that of composite modules, in which a module is itself a smaller workflow. Composite modules are an important mechanism for abstraction, privacy, and reuse [4] between workflows. The idea in this paper is to use composite modules as an abstraction mechanism, driven by user-input on what is relevant for provenance.
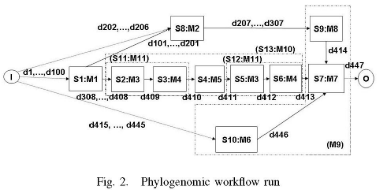
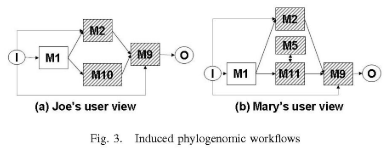
In this paper, we have two goals. First, we want to help users construct relevant user views. For example, Joe would be presented with Ml,.., M8 and indicate that he finds M2, M3 and M7 to be relevant. Based on this input, the user view in Figure 3(a) would be created.
Second, we want to design a system in which the answer to a provenance query depends on the level at which the user can see the workflow. For example, based on the execution in Figure 2, the answer to a query by Mary on what data objects were used to produce d413 would include the data passed between executions of Ml 1 and M5, d410 and d41 1. However, this data would not be visible to Joe since it is internal to the execution of MIO. Thus the answer to a provenance query depends on the user view. Our approach should also be able to switch between user views. While several workflow systems are able to answer provenance queries [5], none take user views into account.
“We call the provenance of a data object the sequence of modules and input data objects on which it depends [5], [6]. If the data is a parameter or was input to the workflow execution by a user, its provenance is whatever metadata information is recorded, e.g. who input the data and the time at which the input occurred. Following other work (e.g. [7], [1]), we assume data is never overwritten or updated in place. Each data object therefore has a unique identifier and is produced by at most one step.”
Contributions. In this paper we propose a model for querying and reasoning about provenance through user views (Section 2). We then define properties of a "good" user view, and present an algorithm which takes as input a workflow specification and a set of relevant modules, and constructs a good user view (Section 3).
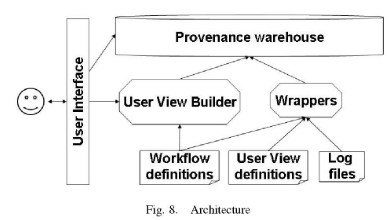
Based on this model, we have built a provenance reasoning system which assists in the construction of good user views, stores provenance in an Oracle warehouse, and provides a user interface for querying and visualizing provenance with respect to a user view (Section 4).
To evaluate our provenance reasoning system, we have created an extensive suite of simulated scientific workflow specifications based on patterns observed in 30 actual work- flows collected from scientists. Measurements include the cost of querying the warehouse, while varying the kind of
workflow, run, and user view (Section 5).
[Unspecified.]
[Formalization of] the notion of user views, [demonstration of] how they can be used in provenance queries, and [...] an algorithm for generating a user view based on which tasks are relevant for the user.
[Description of a] prototype and [...] performance results.
Full results omitted; see full text.
Chebotko, Artem; Lu, Shiyong; Fei, Xubo; Fotouhi, Farshad
RDFPROV
A relational RDF store for querying and managing scientific workflow provenance
2010
[Unspecified.]
Data & Knowledge Engineering
Keywords:
Provenance
Scientific workflow Metadata management Ontology
RDF
OWL
SPARQL-to-SQL translation Query optimization RDF store
RDBMS
Artem Chebotko a,⁎, Shiyong Lu b, Xubo Fei b, Farshad Fotouhi b
a Department of Computer Science, University of Texas-Pan American, 1201 West University Drive, Edinburg, TX 78539, USA b Department of Computer Science, Wayne State University, 431 State Hall, 5143 Cass Avenue, Detroit, MI 48202, USA
In this paper, we propose an approach to provenance management that seamlessly integrates the interoperability, extensibility, and inference advantages ofSemantic Web technologies with the storage and querying power ofan RDBMS to meet the emerging requirements of scientific workflow provenance management. Our motivation of using the mature relational database technology is provided by the fact that provenance metadata growth rate is potentially very high since provenance is generated automatically for every scientific experiment. On the Semantic Web, large volumes ofRDF data are managed with the so called RDF stores, and majority ofthem, including Jena [118,119], Sesame [23], 3store [56,57], KAON [107], RStar [71], OpenLink Virtuoso [42], DLDB [81], RDFSuite [9,105], DBOWL [77], PARKA [101], and RDFBroker [100], use an RDBMS as a backend to manage RDF data. Although a general-purpose relational RDF store (see [15] for a survey) can be used for provenance metadata management, the following provenance-specific requirements bring about several optimization strategies for schema design, data mapping, and query mapping, enabling us to develop a provenance metadata management system that is more efficient and flexible than one that is simply based on an existing RDF store.
• As provenance metadata is generated incrementally, each time a scientific workflow executes, provenance systems should emphasize optimizations for efficient incremental data mapping. As we show in this work, one of such optimizations, a joinelimination optimization strategy, can be developed for provenance based on the property that workflow definition metadata is generated before workflow execution metadata.
• As the performance for provenance storage and that for provenance querying are often conflicting, it may be preferable for a provenance management system to trade data ingest performance for query performance. For example, for long-running scientific workflows, trading data ingest performance for query performance might be a good strategy.
• The identification of common provenance queries has the potential to lead to an optimized database schema design to support efficient provenance browsing, visualization, and analysis.
• Update and delete are not the concern of provenance management since it works in an append fashion, similarly to log management. Therefore, we can apply some denormalization and redundancy strategies for database schema design, leading to improved query performance.
These provenance-specific metadata properties cannot be assumed by a general-purpose RDF store, hampering several interesting data management optimizations to gain better performance for data ingest and querying. While conducting a case study for a real-life scientific workflow in the biological simulation field (see Section 7 for detailed information) to illustrate and verify the validity of our research, we observed that two popular general-purpose RDF stores, Jena and Sesame, could not completely satisfy the provenance management requirements of the workflow. While Sesame could not keep up with the data ingest rate, Jena could not do as good as Sesame on query performance. Both systems lacked support for some provenance queries.
Therefore, by exploiting the above provenance characteristics, we design a relational RDF store, called RDFPROV, which is optimized for scientific workflow provenance querying and management. RDFPROV has a three-layer architecture (see Fig. 1) that complies with the architectural requirements defined for the reference architecture for scientific workflow management systems [68]. The provenance model layer is responsible for managing provenance ontologies and rule-based inference to augment to-bestored RDF datasets with new triples. The model mapping layer employs three mappings: (1) schema mapping to generate a relational database schema based on a provenance ontology, (2) data mapping to map RDF triples to relational tuples, and (3) query mapping to translate RDF queries expressed in the SPARQL language into relational queries expressed in the SQL language. These mappings bridge the provenance model layer and the relational model layer, where the latter is represented by a relational database management system that serves as an efficient relational provenance storage backend.
“This support is enabled via provenance metadata that captures the origin and derivation history of a data product, including the original data sources, intermediate data products, and the steps that were applied to produce the data product.”
The rest ofthe paper is organized as follows. Section 2 reviews related work. Section 3 discusses the provenance model layer of RDFPROV and introduces a sample provenance ontology. Sections 4, 5, and 6 present the model mapping layer of RDFPROV, elaborating on provenance ontology to database schema mapping, provenance metadata to relational data mapping, and SPARQLto-SQL query translation, respectively. Section 7 provides our case study for provenance management in a real-life scientific workflow from the biological simulations field. Section 8 empirically compares RDFPROV with two commercial relational RDF stores. Finally, Section 9 concludes the paper and discusses possible future work directions.
[Unspecified.]
i) two schema mapping algorithms to map an OWL provenance ontology to a relational database schema that is optimized for common provenance queries; ii) three efficient data mapping algorithms to map provenance RDF metadata to relational data according to the generated relational database schema, and iii) a schema-independent SPARQL-to-SQL translation algorithm that is optimized on-the-flyby using the type information ofan instance available from the input provenance ontology and the statistics of the sizes of the tables in the database. Experimental results are presented to show that our algorithms are efficient and scalable.
Full results omitted; see full text.