Citation, Title
Setting
Objective
Methods
Outcome
Results
Conclusion, Notes
(Miles et al. 2011), PrIMe: A Methodology for Developing Provenance-Aware Applications
This article is related to a provenance research project leading to a new approach that is independent of implementation technology or domain.
The authors expand on a less detailed presentation of their PrIMe method which was already given in 2006, at the 6th international workshop on software engineering and middleware.
The presented PrIMe software engineering method aims to make various applications provenance aware, i.e. enable the recording of useful process documentation.
Other goals are ease of PrIMe’s use and the ability to trace back steps taken to use case requirements.
The authors describe the provenance of an entity as the documented record of past events, actions or processes related to this entity. More specifically, the provenance of a data item is defined as the process (series of actions) that led to that item.
The data model used (described by Groth et al. 2009), called provenance architecture, follows the service-oriented style, i.e. state is only created by actors communicating by message passing. Applying PrIMe to a process enables its integration with the architecture – this may require reshaping the process to fit the actor model. The actor model includes interactions between actors, relationships between incoming and outgoing data; and internal actor state.
A provenance store is used to record and query process documentation which consists of p-assertions (about the given process) at specific points in time, stored only by actors with access to the relevant information.
Use cases, called provenance questions, are employed as an analysis tool to find the data most relevant to a given application.
A bioinformatics application is used as a case study throughout the discussion of the model’s steps (the Amino acid Compressibility Experiment – ACE).
Natural-language definitions, non-standard diagrams, XML data structures, and a pseudocode algorithm are used as well.
To show the benefits of the approach, it is briefly compared with an existing, aspect-oriented solution (Jacobson and Ng, 2004).
A method to make applications provenance aware, with the description of processes, methods and structures used to achieve that goal.
An example based evaluation of feasibility and usability; an analysis of traceability and future proofing.
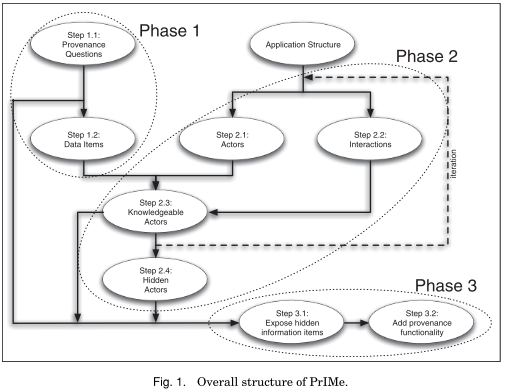
The structure of PrIMe is shown in the graph below. Nodes correspond to method steps and directed edges show their succession. Dashed ovals show three phases, namely:
- Give use cases (provenance questions) to find relevant data and the kind of provenance to be stored for it (scope).
- Decompose the application (into actors and their interactions) to reveal the flow of information. Find the knowledgeable actors with access to provenance information. If necessary, repeat this phase at finer granularity to access missing data items. Note actors that are not adaptable to record provenance info.
- Adapt the application for provenance recording to finally be able to answer the initially posed provenance questions.
Note: the actor model mentioned in the methods column results from applying the methodology shown above to a specific application in order to make it provenance aware.
The recorded process documentation constitutes a second view – showing the application’s execution; a directed acyclic graph (DAG) of the data’s causal dependencies.
Comparing PrIMe to an existing – aspect oriented – approach the authors argue that its main advantage comes from the assumptions it makes, importantly that provenance data can be connected by causal relationships so that once answered provenance questions can be reused in the answer to other questions.
While the authors acknowledge the difficulty of evaluating methodologies, PrIMe is evaluated in terms of usability (satisfied by design, according to the authors), applicability (existing example applications), traceability (back to use cases; shown mainly by means of a decomposition algorithm), and future proofing (partially fulfilled; the authors argue that all provenance questions related to documented processes can be answered). Evaluation metrics are proposed, but not applied, for the designs and implementations resulting from the method.
“As shown in our comparison with an aspect-oriented methodology, PrIMe crystallizes issues that will arise in any project to build a provenance-aware system.”
Note:
Luc Moreau is listed as editor of the W3C PROV data model (current version from April 2013) while Paul Groth and Simon Miles are listed as contributor[1].
(Almeida et al. 2016), A provenance model based on declarative specifications for intensive data analyses in hemotherapy information systems
This paper is an interdisciplinary effort involving researchers from the university of São Paulo and the São Paulo Blood Center (Fundação Pró Sangue – FPS), the largest blood center in latin america (processing ~100,000 whole blood units per year).
The goal is to improve the data quality of a blood donation database, maintained by the São Paulo Blood Center, with the help of a provenance model. Better data quality, in turn, should allow to answer two main questions, namely (1) which donors are at risk of iron deficiency anemia, and (2) what donation number/interval increases that risk. Answering these questions should help to reduce undesirable outcomes for blood donors while the provenance data should enable understanding, control and reproducibility of each step of the process.
The authors mention a definition of provenance as documentation of the history of data, including each transformation step.
In order to answer questions about blood donor anemia, the authors determine relevant donor groups in a step-by-step selection- and distinction process by applying (and emulating) specialist human knowledge to a large dataset. For example, FPS researcher’s knowledge was used to define invalid data values. Even though not explicitly stated, it seems likely that most selection criteria used in the derivation of groups are based on export knowledge.
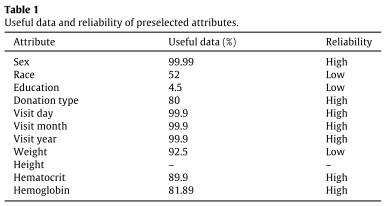
In preprocessing, suitable attributes for analysis were selected according to usefulness (>80% valid entries), high reliability (objectivity), and donor age (from 18 to 69); normalization to Hct (hematocrit) values was performed; Brazilian National Health Surveillance Agency cut-off values for Hb (hemoglobin) and Hct were used.
Each step of the model was evaluated with respect to the desired categories, both statistically and based on data annotation and validation performed by experts (no details given).
The data resulting from applying the whole model underwent a preliminary descriptive statistical analysis based on mean plots, a life table estimator and multivariate analysis based on a logistic (due to binary variable) mixed (due to possible dependence among donations) model to exemplify suitability and reliability of the generated groups.
A question-based model of successively refined (blood donor) data groups designed to understand the reasons for deferrals due to low Hct in regular blood donors.
Use-case based evaluation of the generated groups (statistical analysis).
The result of classifying useful and reliable blood donor attributes (cf. methods) for analysis, with the help of experts, is shown in Table 1.
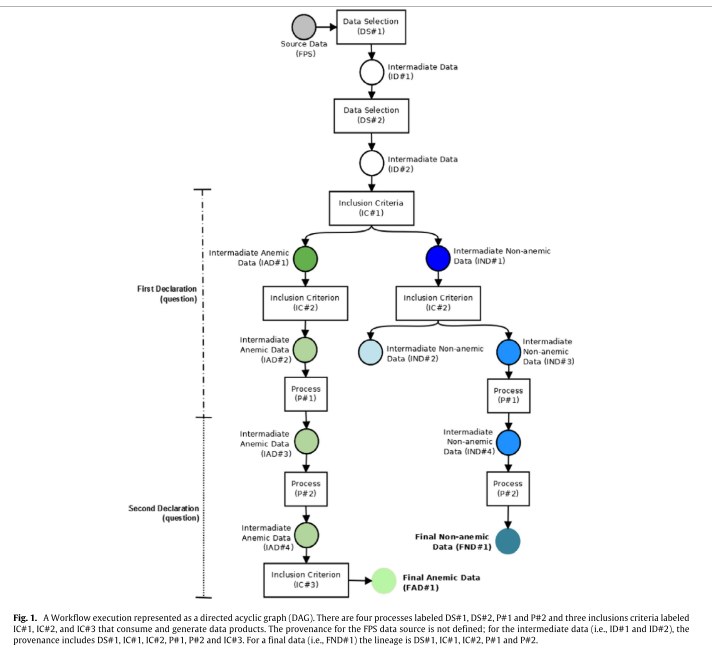
To create the study groups used in the statistical analyses, the rules (inclusion criteria) and transformations (filters) shown in the workflow of figure 1 were used, which illustrates the provenance of the intermediate and final data.
Their functions are the following: DS#1 selects records for the period studied (from 1996 to 2006), DS# 2 cleans the dataset (cf. methods), IC#1 classifies blood donors according to their risk of developing anemia. The second inclusion criterion, IC#2, separates the donor groups according to the number of donations performed. The resulting subgroups are non-anemic first-time donors (IND#2) non-anemic two-or-more-times donors (IND#3 and IND#4) and anemic (former) regular donors (IAD#2 and IAD#3). Then, P#1 normalizes the records (Hb to Hct), P#2 inserts donation intervals and the number of blood donations (both used in the analysis) into the dataset and, finally, IC#3 selects only the dataset of anemic regular donors (IAD#4) that were not deferred due to low Hct in the first recorded donation.
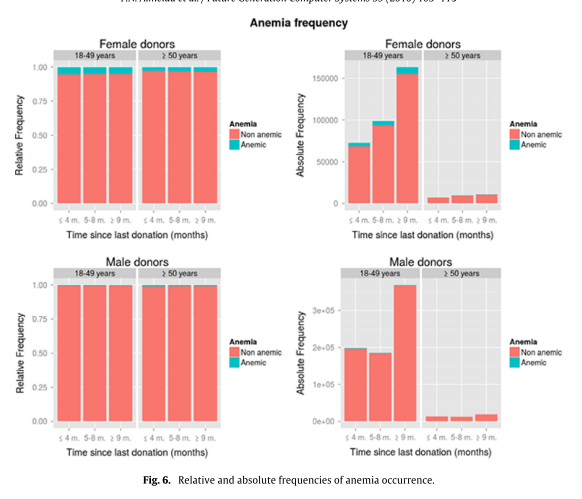
Results show that the probability of anemia after a donation is inversely related to previous Hct levels for both men and women. However, young and female donors are especially at risk of developing anemia, and developing it earlier.
The use-case based evaluation includes statistical analyses based on the obtained study groups, namely a life table estimator, mean Hct values and donation intervals, as well as relative and absolute frequencies of anemia occurrence (for groups of gender, age and time since last donation – in the latter analysis, shown below).
It seems apparent that young and female donors are especially at risk of developing anemia.
The feasibility of the model with respect to the usefulness of the generated groups is shown by means of the above use case. However, no comparison is made with existing models, and no formal measure of fitness is given.
“This paper shows that the provenance model is important for the generation of a reduced dataset and to provide efficient and conclusive analyses based on expert knowledge. The provenance was used to extract and select knowledge from a huge data volume.”
(Amanqui et al. 2016), A Model of Provenance Applied to Biodiversity Datasets
Researchers from Ghent University’s Data Science Lab and University of Sao Paulo’s ICMC contributed to this article in the context of biodiversity research.
The goal is a conceptual provenance model for species identification. A primary requirement is interoperability in heterogeneous environments such as the web.
The model’s evaluation is performed on a use case basis.
The authors describe provenance in the context of species identification as the history of the species – meaning the process of identification – which typically involves different persons possibly far apart in space-time.
Five biodiversity scientists were asked to identify key biodiversity information like what was collected, where, how and by whom. The information was gathered in the form of biodiversity use cases obtained from structured interviews[2] with questions mostly asking for user, goal, motivation, tasks and desired tool features. A typical species identification process is illustrated below. The use cases are employed to create and evaluate the model (cf. results, figure 3).
W3C PROV is used as the basis for the model[3][4].
A mapping from ontologies, provenance model, taxonomy and collection database to RDF (using the RML language) is defined for, and OWL is used to represent, the biodiversity data. RDF enables queries based on relationships using SPARQL. The mapping consist of Logical Source (dataset), Subject Map (resource URIs) and optional Predicate-Object Maps.
GeoSPARQL ontology terms and Well-Known Text (WKT) are used to describe georeferenced data.
The ProvValidator and ProvTranslator web services are used to provide data provenance validation and interoperability, respectively.
A conceptual model for provenance in biodiversity data for species identification.
The model’s feasibility is demonstrated by applying it to a use case.
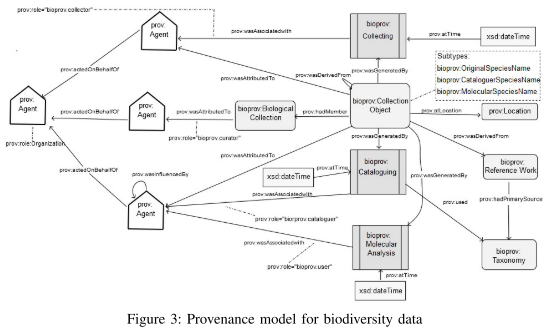
The resulting provenance model is shown below in figure 3, as a graph. The nodes fall into the three (W3C PROV) classes Entity, Agent and Activity. Edges connecting these classes form relations between them.
To model the (re)naming of species by – possibly different – agents, the authors extend the W3C PROV model by the following three subtypes of prov:Entity:
- bioprov:OriginalSpeciesName is given to a species by the collector at the very start of identification.
- bioprov:CataloguerSpeciesName is derived from an original name by the cataloguer.
- bioprov:MolecularSpeciesName results from changing an existing species name.
The extended model is available online[5].
As a demonstration of feasibility, the model was applied to the use case of genetically identifying the alga species Cladophora Delicatula (cf. methods, interview 1). A selection of the resulting definitions is shown below (the full provenance (ttl) is available online[6]).
:AgentCollector a prov:Agent; foaf:givenName "M.C.Marino & R.Marino"; prov:actedOnBehalfOf :IBT.
:Collecting a prov:Activity; prov:wasAssociatedWith :AgentCollector; prov:atTime "1966-11-11T01:01:01Z"; bioprov:OriginalSpeciesName Cladophor Delicatula.
:Location a prov:Entity; foaf:name "Ponta do Gil Lake"; prov:qualifiedGeneration [ a prov:Generation; prov:activity geo:feature; prov:atTime "1966-11-11T01:01:01Z"; prov:atLocation "POINT(-46.7175 -23.653056)"ˆˆgeo:wktLiteral;]
:AgentCataloguer a prov:Agent; foaf:givenName "D.P. Santos"; prov:actedOnBehalfOf :IBT .
:Cataloguing a prov:Activity; prov:wasAssociatedWith :AgentCataloguer; prov:atTime "1982-01-01T01:01:01Z"; bioprov:CataloguerSpeciesName Cladophor Delicatula
Applying their methods the authors were able to query their RDF data for the lineage of Cladophora Delicatula. No precision/recall evaluation of the queries is performed, although planned for later work.
“In this work, we presented a model for biodiversity data provenance (BioProv). BioProv enables applications that analyze biodiversity to incorporate provenance data in their information. We use the provenance information to allow experts in biodiversity to perform queries and answer scientific questions.”
Notes:
No precision/recall evaluation of the queries is performed, although planned for later work.
(Groth, Miles, and Moreau 2009), A Model of Process Documentation to Determine Provenance in Mash-Ups
The article’s authors previously published on architecture, performance and uses of provenance systems. The data model used in these systems is explicitly described in this article, along with its foundations.
A conceptual process documentation model that enables the creation of documentation independently of the number of involved software parts, institutions and application domains – and applying the documentation to find the provenance of obtained results.
The model is evaluated by means of a bioinformatics use case.
The authors define the provenance of a result as the process which led to that result.
Nonfunctional requirements (derived from use cases, in the authors’ previous work) are stated which the model should enable, namely that process documentation has to be
- factual,
- attributable, and
- autonomously creatable.
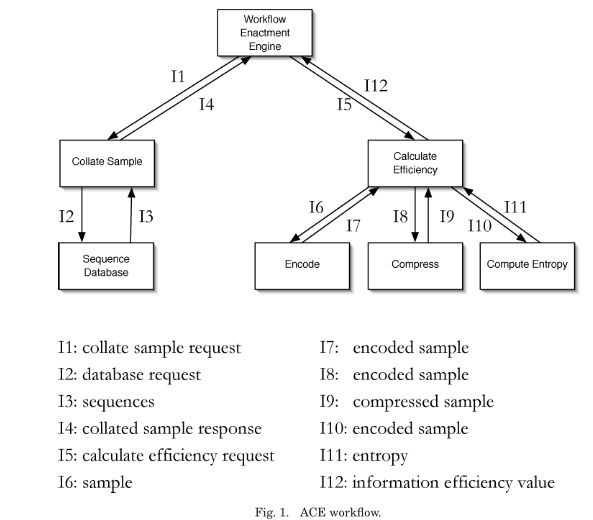
The Amino acid Compressibility Experiment (ACE) serves as an example mash-up use case for presentation and evaluation of the proposed model, named p-structure. A workflow of the experiment is shown below.
Concept maps are used to provide a human-friendly overview of the concepts and the relationships between them. Computer parsable representations are provided in the form of XML and OWL [7].
An actor-centric view of applications is adopted, where each actor component represents a set of functionality and interacts with other actors by sending and receiving messages through well-defined communication endpoints.
A qualitative evaluation of the data model with respect to the above requirements and the use case’s provenance questions is given.
The actor Collate Sample is implemented in Java as a web service. Compress and Calculate Efficiency are run as jobs on a grid.
The PReServ provenance store is used for tests; it offers XQuery and a provenance specific graph traversal query function. Using these query mechanisms, the use case questions are answered.
Firstly, the authors provide a generic, conceptual data model for process documentation.
Secondly, the feasibility of the model is evaluated with the help of a bioinformatics use case.
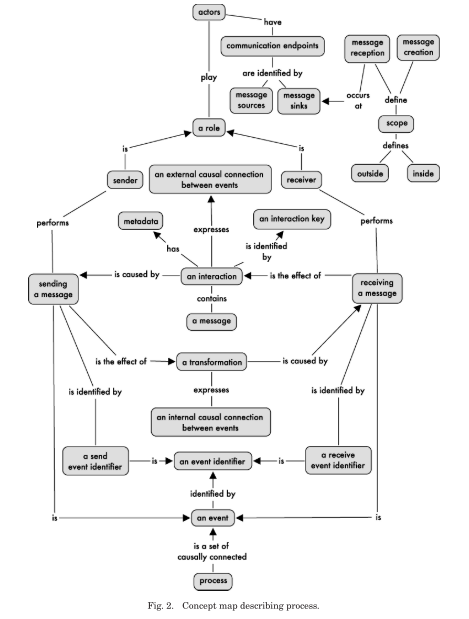
The authors introduce p-assertions, which contain the elements necessary to represent a process. Below is the authors’ concept map description of a process. In such a map, relationships between concepts are read downwards or, if an arrow is present, in the arrow’s direction.
To satisfy the requirement of factuality, a restriction is placed on all asserters (actors) that they only create p-assertions for events and data that are directly accessible by them. P-assertions also have an asserter identity, satisfying the requirement of attributability.
Interactions are represented by interaction p-assertions, which contain
- asserter identity,
- event identifier,
- message representation, and
- description of how the representation was generated.
A relationship p-assertion identifies at least one cause and, for simplicity, only one related effect. This kind of p-assertion documents both the data flow and control flow within an actor and is thus critical to understanding the process within an application.
While interaction and relationship p-assertions are sufficient to document a process, another kind, namely the internal information p-assertion, is introduced, which models message reception, and can be used to abstract away details of how data items come about. Its contents are similar to an interaction p-assertion and lack the asserter identity.
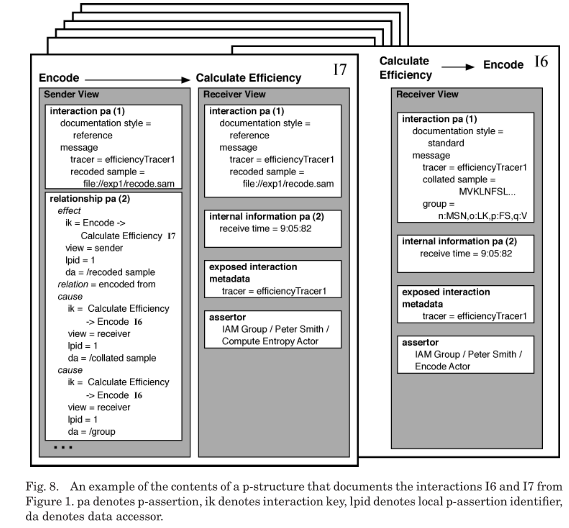
The figure below shows how p-assertions are are grouped together in a view by a common event to form p-structures. P-structures are an organizational tool for finding and understanding groups of p-assertions.
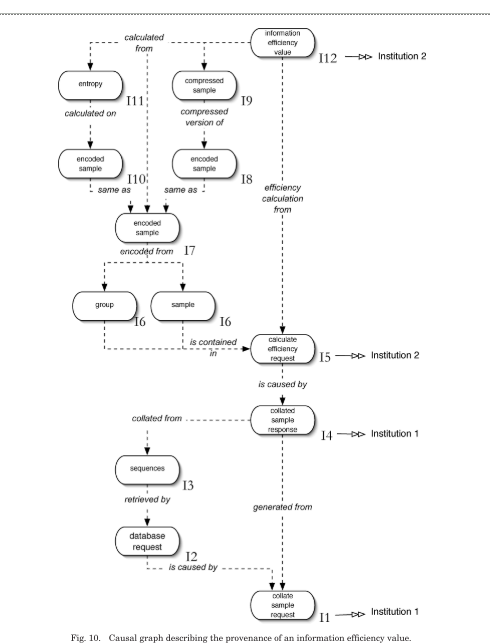
To describe the provenance of a particular event occurrence, a causal graph whose connections lead back to that event can be created from the p-structure:
The nodes in the graph are occurrences, in the role of causes, effects or both. The edges in the graph are hyper-edges and represent the causal connections extracted from relationship p-assertions. All arrows on the edges point from effect to cause.
The external causal connections represented by interaction p-assertions are collapsed into the numbers shown to the bottom right of each node.
In the evaluation of their model, the authors show how it satisfies the previously stated requirements (cf. methods section), namely by design of the p-structure, also discussed above, as long as users adhere to the specification.
Also shown is how to answer the four ACE use case questions using a causality graph created from the process documentation.
“We have defined the concepts that comprise a generic data model for process documentation, the p-structure. This description included several concept maps that unambiguously summarize the various concepts underpinning the data model and the relationships between them. The data
model supports the autonomous creation of factual, attributable process documentation by separate, distributed application components.”
Note:
Luc Moreau is listed as editor of the W3C PROV data model (current version from April 2013) while Paul Groth and Simon Miles are listed as contributors (“PROV-DM: The PROV Data Model” 2017).
(Ma et al. 2015), Causal dependencies of provenance data in healthcare environment
Researchers from Chinese (University of Hebei, Taiyuan Nonnal University) and Australian (Victoria University, University of Southern Queensland) institutions contributed to this article in the context of provenance in healthcare.
In order to support features like access control, the goal is to explicitly include the causal dependencies in a provenance model.
The authors describe provenance as the history of data, or information about the origin, derivation, describing how an object came to be in its present. And, specifically, the provenance of a data object as the documented history of the actors, communication, environment, access control and other user preferences that led to that data object.
The Open Provenance Model (Moreau et al. 2011) is used as a basis which the authors seek to enhance.
Model development is based on a list of requirements (from previous work), namely the model has to
- be fine grained
- provide provenance security
- support various types of provenance queries and views
Provenance records are represented by a relational DBMS.
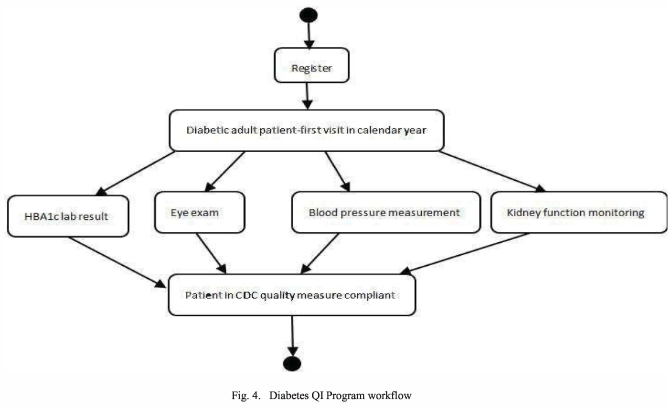
The model’s feasibility is assessed by a use case: applying it to represent an actual healthcare workflow (from a – not otherwise specified – Diabetes Quality Improvement Program)
An open provenance model for access control which explicitly captures causal dependencies.
A use case to show the feasibility of the proposed model.
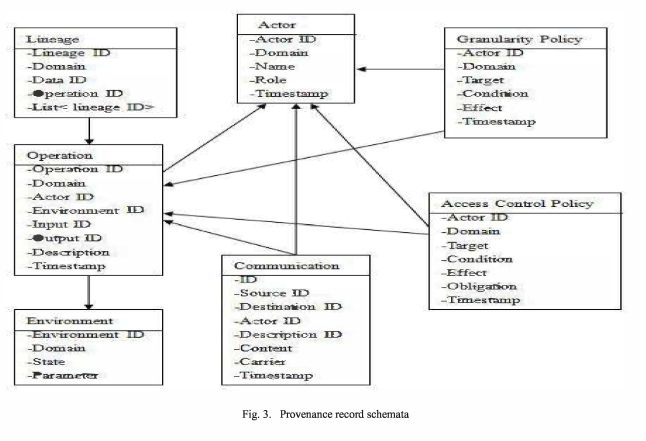
The proposed provenance (access control) model is shown below. It is based on the Open Provenance Model (Moreau et al. 2011) and contains the entities that are part of the definition of the provenance of a data object (cf. methods), and shows the interactions among them.
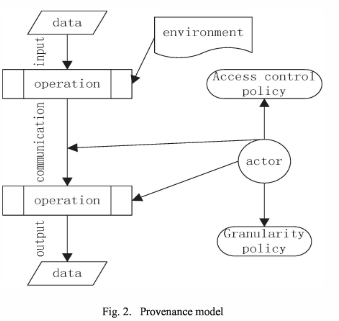
Central to the model are actors, which carry out operations on data, and communicate the data between different operations. Data arises and changes because of (a sequence of) operations. Each datum is associated with a record of the data objects that have been directly used in its creation. This may also be data in the environment, that is not explicitly part of an operation’s input but affects its output. To provide access control for an operation, the model includes an access control policy and a granularity policy. The access control policies, like other parts of the model, are also directed by actors.
The schematic provenance records for the various model entities are shown below, each of them uniquely identified. With the directed edges between them they form a provenance DAG (directed acyclic graph).
The following is a short description of the use case application of the proposed model.
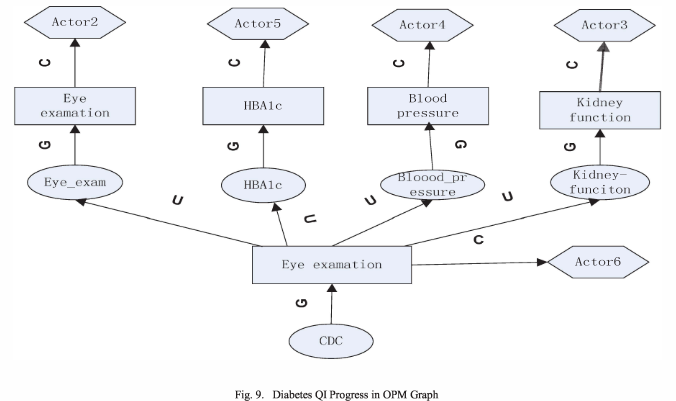
The authors proceed to apply the proposed provenance model to the medical records generated from a Diabetes Quality Improvement Program workflow, shown below, which enables them to capture the provenance of this process (details omitted here) and shows the model’s feasibility.
The authors also use the OPM model to determine the causal dependencies, shown below. In the figure, artifacts (data objects) are represented by ellipses, processes (operations) are represented by rectangles, agents correspond to actors and the environment. C, U, and G are sets of role-specific variations of 'was controlled by', 'used', and 'was generated by' (causal relationships of the OPM model).
“We apply the OPM model to access control the provenance information. So it can explicitly capture the causal dependencies where the access control policies and other preference based on” [sic]
(Schreiber 2016), A Provenance Model for Quantified Self Data
The article was written in the Distributed Systems and Component Software group at the German Aerospace Center (DLR) in Cologne.
As part of the broader vision to understand the creation of – and enable security and trust in – QS (quantified self) data with the use of provenance, the author proposes an ontology and a provenance data model for this use case.
Such a model specifies, in a standardized way, what data will be stored, and can thus be instrumental in solving the suggested problems.
The ontology is meant to enhance the information value of the provenance by providing semantics.
The model’s feasibility shall be evaluated on the basis of a fitness tracker use case.
The author mentions synonyms of provenance from different domains, namely lineage, genealogy and pedigree, and employs the W3C’s definition of provenance (“PROV-DM: The PROV Data Model” 2017) as “a record that describes the people, institutions, entities, and activities involved in producing, influencing, or delivering a piece of data or a thing. In particular, the provenance of information is crucial in deciding whether information is to be trusted, how it should be integrated with other diverse information sources, and how to give credit to its originators when reusing it. In an open and inclusive environment such as the Web, where users find information that is often contradictory or questionable, provenance can help those users to make trust judgments”
Both the provided ontology and provenance model are based on W3C standards, namely PROV-O (“PROV-O: The PROV Ontology” 2017) and PROV-DM (“PROV-DM: The PROV Data Model” 2017), respectively, where the ontology was created based on an ontology development guide by Noy and McGuiness[8] and the provenance model based on analyzing requirements of quantified self workflows and documentation created at conferences.
Formal modelling methods are – apparently – used, namely the Protégé ontology editor[9] and description logics (“OWL Web Ontology Language Reference” 2017).
To focus the recording of provenance on the process parts relevant to the problem at hand, the author poses 10 questions about a user’s quantified self data, split between the basic constituents of the provenance model (entity, activity and agent).
The feasibility of the presented provenance model is shown by means of a workflow using steps data from the Fitbit fitness tracker.
An ontology and a provenance data model (descriptive and partly formalised) for quantified self workflows.
Use case based evaluation, visualizing Fitbit fitness tracker data (informal).
The author’s solution to the problem of analyzing and securing QS (quantified self) data created by devices such as fitness trackers involves an ontology and a provenance data model for this kind of data.
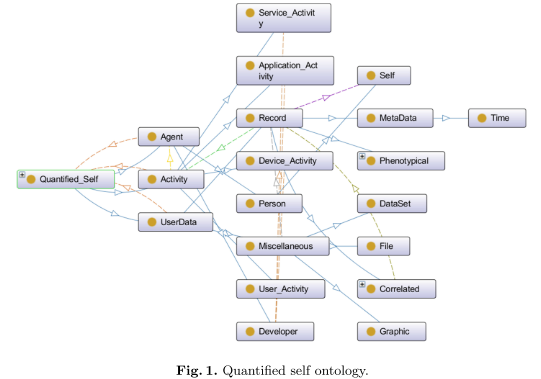
The ontology, pictured below, corresponds to the W3C’s PROV-DM, where Agents and Activities have the same name, and Entities are named UserData.
The PROV-DM classes were extended by a (QS-)Developer (Agent subclass), Self (the user, a Person), as well as User, Device, Application and Service Activities and UserData, Miscellaneous (data) and Record (raw data) Entities.
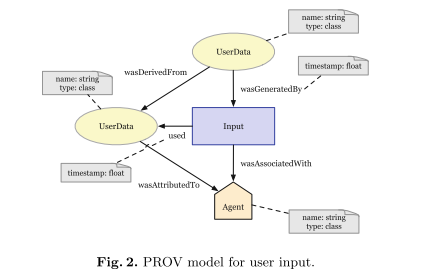
For each QS workflow activity (derived from requirement analysis) one provenance model was created, namely for Input (structure raw user input), Export, Request (data from web/cloud services), Aggregation (helping to interpret data from multiple sources) and Visualization activities. The model for user input is shown below exemplifying the basic building blocks.
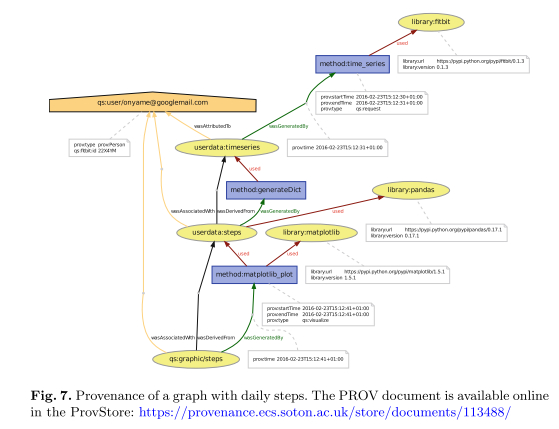
To show the feasibility of the proposed models, the author provides a use case involving the cleaned provenance recording, and visualization of data from the Fitbit fitness tracker. The full example, implemented in Python, is available online[10]. The resulting provenance visualization shows how Fitbit data is used to create a graphical representation of steps walked by the user:
The model functions, according to qualitative evaluation.
“Using that model, developers and users can record provenance of Quantified Self apps and services with a standardized notation. We show the feasibility of the presented provenance model with a small workflow using steps data from Fitbit fitness tracker.”
(Zhao et al. 2004), Using semantic web technologies for representing e-Science provenance
The research presented in this article is part of the U.K. e-Science project myGrid, which is automating in silico experiments on grids – including management of the results.
The authors collaborated with Manchester University biologists and bioinformaticians to research the genetic basis of Williams-Beuren syndrome.
The broader goal is to satisfy the requirements of life science with existing semantic web tools.
This article specifically seeks to enhance provenance data by semantic web tools, e.g. RDF and ontologies.
The authors describe provenance (of an experiment) as records of where, how and why results were generated, and provenance within e-Science describes from where something has arisen.
e-Scientists’ requirements for provenance are stated (apparently from the author's own experience).
RDF and ontologies are used to enhance provenance representation.
LSID and Jena are used for provenance data creation and storage.
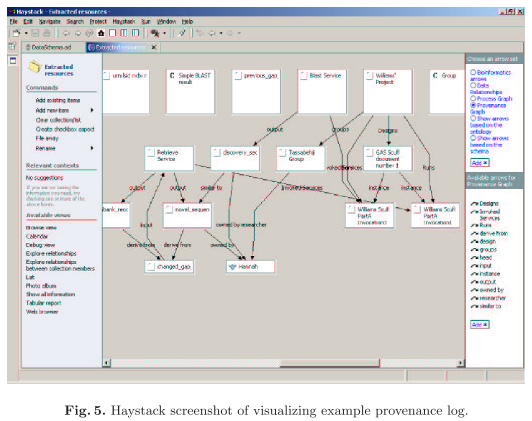
Haystack is used for visualisation, creating multiple views of the data.
A case study exploring the feasibility of using semantic web technology to enhance provenance data for the benefit of e-Scientists.
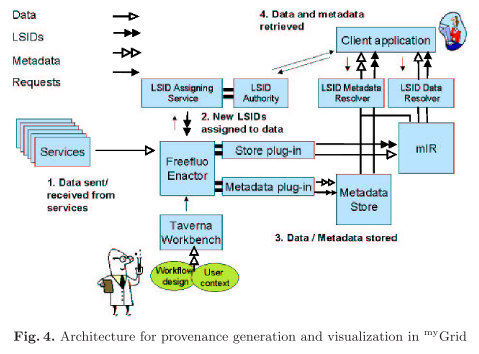
The authors present the provenance architecture pictured below.
Experiment workflow provenance is represented by RDF and identified by LSIDs. Four views on these data are recorded in myGrid, namely process, data derivation, organisation, and knowledge (semantic) views. Process and data derivation provenance are automatically captured; selected organisational provenance is added to experimental data, and knowledge provenance is added by means of Taverna (a workflow environment) templates and the myGrid bioinformatics services ontology. The resulting provenance data is visualized with Haystack:
The authors recognize that scalability – as far as provenance processing and presentation are concerned – is yet to be assessed.
“Given our addition of a semantic description of these data, we fully expect to
be able to add machine processing of these data to the current human emphasis
of our work. [...] This would allow
semantic querying of these logs, provide trust information about data based on
logs, etc.”
(Zhong et al. 2013), Developing a Brain Informatics Provenance Model
Researchers from Chinese (Beijing University of Technology, The Hong Kong Polytechnic University, Beijing Key Laboratory of MRI and Brain Informatics), Japanese (Maebashi Institute of Technology) and Dutch (Vrije University Amsterdam) institutions contributed to this article in the context of Brain Informatics.
The goal is to improve modelling accuracy of brain data, as well as integration of data from various sources, by developing a provenance model for brain informatics.
A case study shall show the model’s feasibility and usability.
According to the authors, “provenance information describes the origins and the history of data in its life cycle”.
The Open Provenance Model (Moreau et al. 2011) is used as a basis for – and is extended to – the proposed model.
A case study (in ”thinking-centric systematic investigation”) is used for feasibility/usability assessment.
Firstly, a provenance model for brain informatics data.
Secondly, a case study to show the feasibility and usefulness of the proposed model.
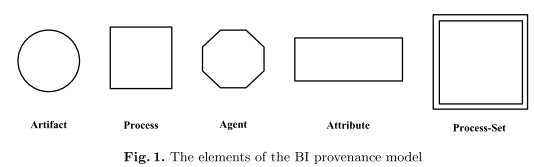
The resulting brain informatics provenance model includes basic and extended types of model elements.
Based on OPM, the three basic elements (with their definitions) are:
- Artifact - an immutable piece of state used or produced during BI experiments or data analysis
- Process - an action or a series of actions … during BI experiments or data analysis
- Agent - contextual entity acting as a catalyst of a process
To satisfy the requirements of brain informatics data (and analysis) provenance modelling to capture the different characteristics of specific basic elements, two extended elements are introduced:
- Attribute - a mapping: At : E → C,S, T,N, or ∅ where E={e | e is an Ar, Pr, or Ag}, C is a set of characters, S is a set of strings, T is a set of texts, and N is a set of numbers, for describing a characteristic of artifacts, processes or agents
- Process-Set - a set of processes: {prs |∃At,At(prsi)= v ∧ prsi is a Pr, i =1... n}, where v is a character, string, text or number
The conceptual elements of the proposed brain provenance model, based on the Open Provenance Model (Moreau et al. 2011) , are shown below (Artifact, Process, Agent, Attribute, Process-Set).
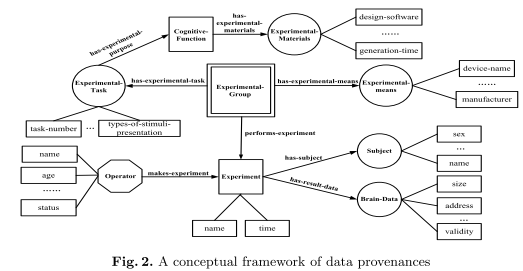
The proposed frameworks of data provenances and analysis provenances, which use the above elements, are shown below.
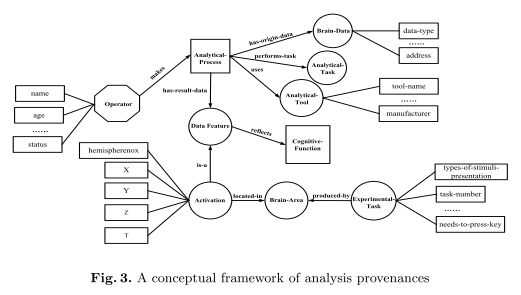
A brain data provenance describes the data’s origin including different aspects of the experiments etc. that produced the data. Figure 2 shows the general conceptual framework for data provenance. Specific datasets need corresponding frameworks with specific elements, i.e. specific artifacts, processes, agents, process-sets and attributes – the same goes for the analysis provenance framework shown in Figure 3.
A brain analysis provenance describes the analytic processing that has been applied to a dataset, e.g. the analysis tasks themselves and their inputs and outputs. A general conceptual framework of analysis provenances is shown in Figure 3.
Specific provenance frameworks are used for the case study in thinking-centric systematic investigations in order to show the feasibility and usability of the proposed model.
“Such a provenance model facilitates more accurate modeling of brain data, including data creation and data processing for integrating various primitive brain data, brain data related information during the systematic Brain Informatics study”.