Results
Following PRISMA, this review’s article inclusion- and exclusion process is depicted as a PRISMA diagram:
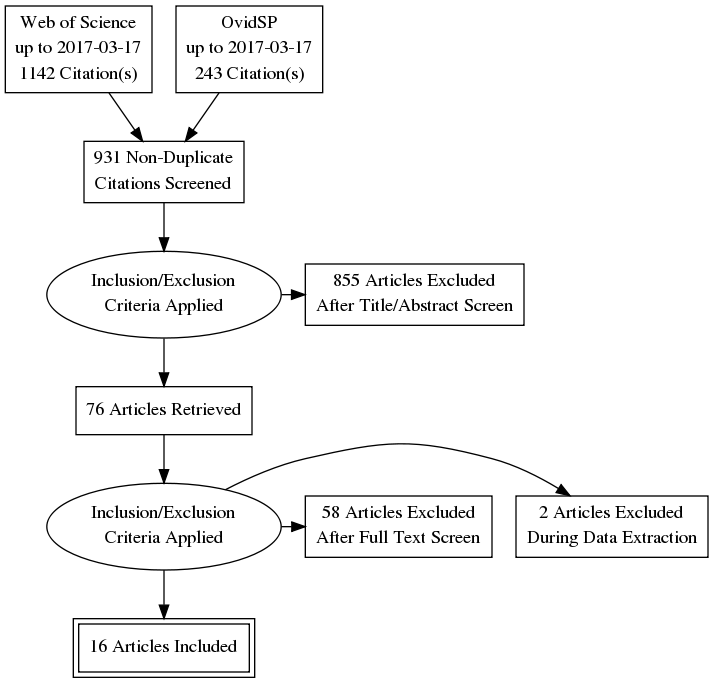
Starting with the articles resulting from searches within Web of Science and OvidSP databases (query: provenance AND medic*; resulting CITAVI database files: Web of Science, OvidSP, merged) the diagram shows each in- and exclusion step as an oval. The numbers of resulting articles are shown in rectangles where the final number of 16 included references represents articles dealing primarily with provenance modelling (7 articles, 2 other articles were excluded during data extraction – a brief review and a very brief letter-style article ), tracking (6) and querying (3, one article recategorised from modelling after data extraction). The 58 references excluded after full text screening include books (4) and articles primarily concerned with datasets (1), data analysis (3), data quality (7), e-health security (5) and -systems (5) and various applications not strictly focused on provenance tracking and querying (33).
Raw- and summary data-sheets
Raw data extraction was performed for all 16 included articles (raw data-sheet).
For the 7 articles focused on provenance modelling, a summary data-sheet was created.
The tabular summary data-sheet should enable the reader to quickly gain an overview of each work as well as the ability to easily compare individual aspects or whole articles.
Because the model presentations used in the original articles are often very different from each other, and use nonstandard diagrams, we found it necessary to unify, and sometimes abstract, them a final table focused solely on the models themselves. To this end we created a standardised representation for each base model, using the textual and graphical information in the summary table.
Models data-sheet
The following models data-sheet bases on the summary data-sheet linked above and contains only the 7 references dealing mostly with provenance modelling. While the summary sheet gives an overview of each whole article, the models data-sheet allows a focused look at the models used in each work.
The models data-sheet's content stems from the following sources in the summary data-sheet – which may have been reworded and clarified. The provenance definition comes from the methods column of the summary table; the model types and representations as well as their relationships and entities mainly come from the results and method columns. We created UML diagrams based on figures or text in the articles and abstracted if necessary.
Next to the article reference, each row contains a provenance definition from the article, because it lays the groundwork for the following models. A unified visual representation of the model or its key aspect shows the model elements, their interaction, and allows comparison between models. The types of models as well as their representations used in the article are made clear. Finally, the base model elements and their relationships are listed (details included in the full, formal model specifications are omitted for efficiency and clarity). This information should give a good overview of the models devised and used in the reviewed works. Model aspects listed separately also facilitate comparison between the approaches.
Zhong et al. 2013 (textual summary)
| Provenance definition | Base model diagram |
|---|---|
|
According to the authors, “provenance information describes the origins and the history of data in its life cycle”. |
|
| Model representations | Objects and dependencies |
|
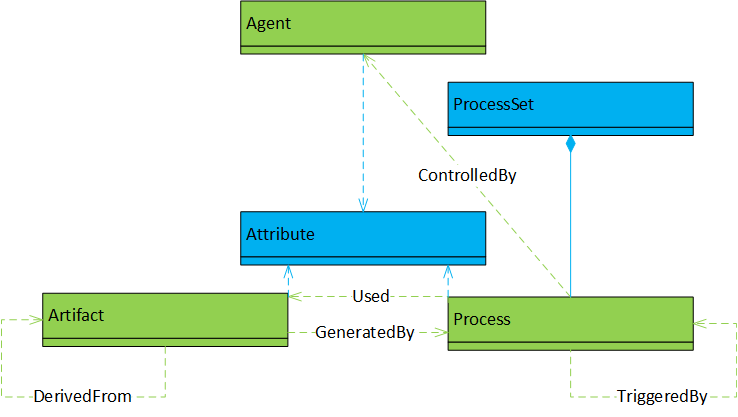
Acyclic graph. |
Objects and dependency relationships correspond to (parts of) the OPM, where objects are (instances of sub-classes of) artifacts, agents, or processes. Additionally, each of the main object instances can have attributes and processes can be combined to process-sets; the OPM’s dependency relationships are:
Additionally, there can be (unnamed) dependencies of agents, artifacts and processes on their attributes. |
Ma et al. 2015 (textual summary)
| Provenance definition | Base model diagram |
|---|---|
|
The authors describe the provenance of a data object as the documented history of the actors, communication, environment, access control and other user preferences that led to that data object. |
|
| Model representations | Objects and dependencies |
|
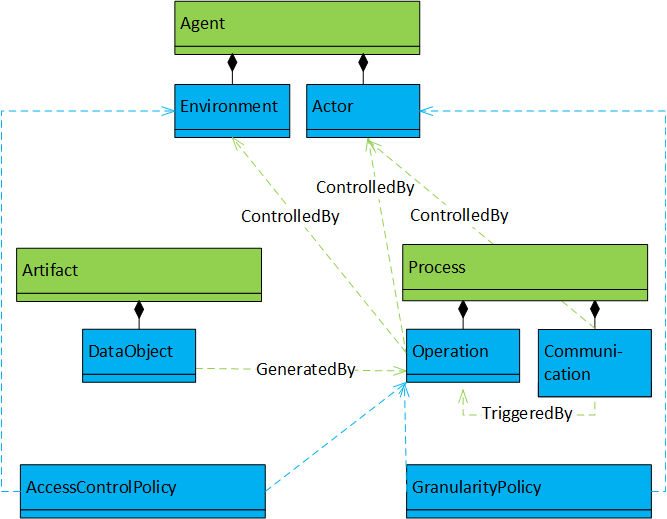
Directed acyclic graph (DAG). |
Objects and dependency relationships correspond to (parts of) the OPM, with additional access-control- and granularity policies. Objects are (instances of sub-classes of) artifacts, agents, or processes; dependency relationships:
Additionally, there can be (unnamed) dependencies of access-control- and granularity-policies on processes or agents. |
Groth, Miles, and Moreau 2009 (textual summary)
| Provenance definition | Base model diagram |
|---|---|
|
The authors define the provenance of a result as the process which led to that result. |
|
| Model representations | Objects and dependencies |
|
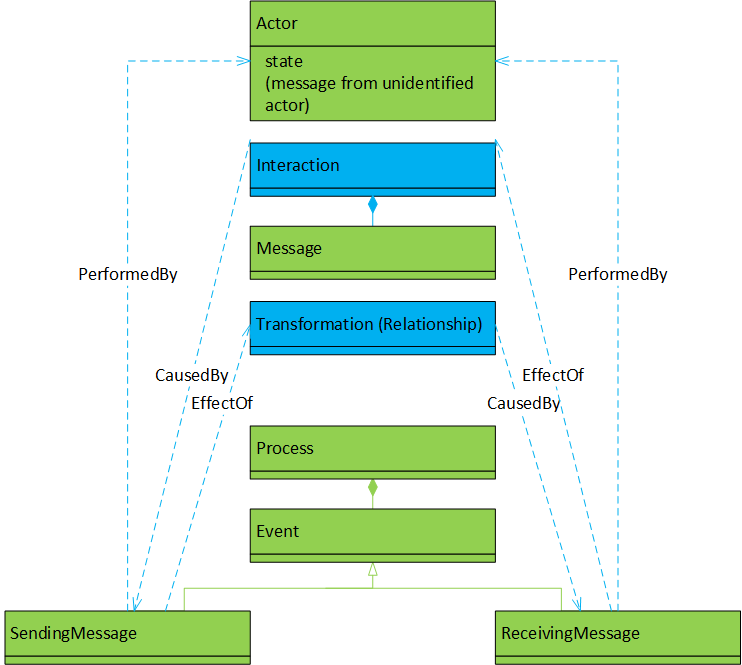
Directed acyclic graph (DAG). The nodes in a provenance-representing graph are occurrences (events and data at an event) in the role of causes, effects or both. The (hyper-)edges in such a graph represent the causal connections. |
Interactions between actors (with internal state) by sending/receiving messages, causal relationships between incoming and outgoing message data. |
Miles et al. 2011 (textual summary)
| Provenance definition | Base model diagram |
|---|---|
|
The authors describe the provenance of a data item as the process that led to that item. |
|
| Model representations | Objects and dependencies |
|
Actor model, leading to a process documentation model (of the same application). The recorded process documentation shows the application’s execution; a directed acyclic graph (DAG) of the data’s causal dependencies. |
Interactions between actors (with internal state) by sending/receiving messages, causal relationships between incoming and outgoing message data. |
Schreiber 2016 (textual summary)
| Provenance definition | Base model diagram |
|---|---|
|
The author cites the W3C’s definition of provenance as “a record that describes the people, institutions, entities, and activities involved in producing, influencing, or delivering a piece of data or a thing [..]” |
|
| Model representations | Objects and dependencies |
|
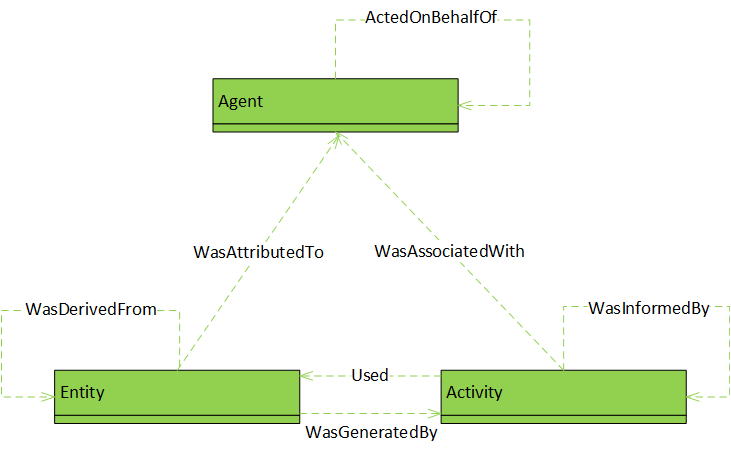
Directed acyclic graph (DAG). |
The same as in W3C’s PROV model. Objects are entities, agents, or activities; dependency relationships:
|
Amanqui et al. 2016 (textual summary)
| Provenance definition | Base model diagram |
|---|---|
|
The authors describe provenance in the context of species identification as the history of the species – meaning the process of identification – which typically involves different persons possibly far apart in space-time. |
|
| Model representations | Objects and dependencies |
|
Directed acyclic graph (DAG). |
Mostly the same as in W3C’s PROV model. Objects are entities (including custom sub-classes), agents, or activities; dependency relationships:
|
Almeida et al. 2016 (textual summary)
| Provenance definition | Base model diagram |
|---|---|
|
The authors mention a definition of provenance as documentation of the history of data, including each transformation step. |
|
| Model representations | Objects and dependencies |
|
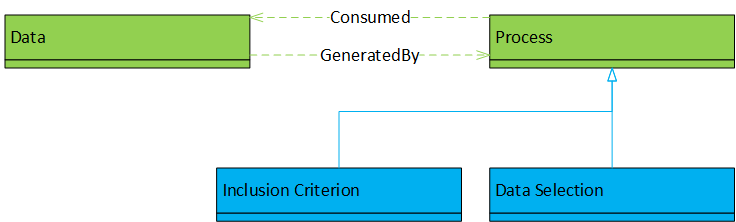
To create study groups for statistical analyses, inclusion criteria and transformation processes were applied to the input data. Provenance information could be obtained for the intermediate and final data. Workflow execution is represented as a directed acyclic graph (DAG). The work shows the creation of a specific model instance for a particular purpose rather than a framework or meta-model, but the approach seems sufficiently general to be applied to similar problems at other institutions. |
Transformation processes, inclusion criteria, data (input, transitional, output); relationships of consumption and generation. |
The standardised models data-sheet enables the conclusion that all but one of the reviewed models are part of the same lineage. We describe the models in the order of that lineage.
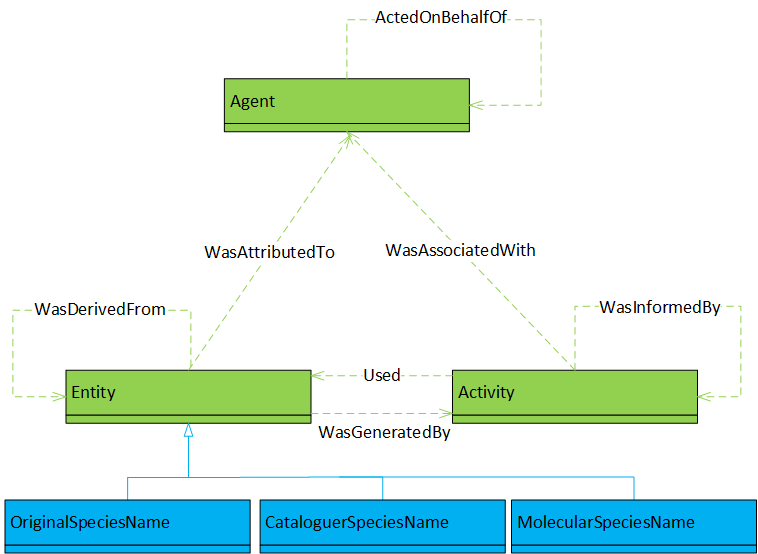
The first two modelling works, and , extend the Open Provenance Model (OPM, ), the lineage’s starting point for our purposes. While the extension is straightforward for , the base model diagram for is more complicated because the authors did not directly base their model on the OPM, and used their own terminology. The diagram illustrates that the model can be represented as an extension of the OPM. In all diagrams, extensions to the reference base model are shown in blue.
The next group of modelling articles contains and . They are special because they fall in between the OPM and its successor, , and form part of ’s development work. As such, these works are simultaneously the most detailed and conceptual of our modelling review. While the OPM’s Agent/Actor and Process types are readily recognised, the Data is more hidden is this transitional model. Since the model incorporates message passing between actors – cf. – as a central concept, the data is implicit in the messages, with the possibility to make it explicit again as internal actor state. On the other hand, the transformative relationship between incoming and outgoing messages (data) is more explicit in this model diagram than in the ones of both OPM and PROV, where it is hidden as the combination of the Used and GeneratedBy dependencies. The final aspect emphasised by this group of models is the relationship between causes and effects – an important aspect in the development of the PROV model but perhaps more implicit than explicit in its use so far.
We end our overview of the models data-sheet with the group of models based on PROV, namely and . The former uses PROV in its plain form, the latter extends it by domain-specific sub-types of the Entity type. We also group the model used by into this category. Even if it is not explicitly based on PROV, the elements of this specific model can be abstracted and then be represented by the more general PROV base model, as done in our diagram.
The similarity between the OPM and PROV base models should be apparent, as their three main types Agent, Artifact/Entity, Process/Activity directly correspond, and the depicted dependencies of PROV are a super-set of those of OPM if we equate TriggeredBy with WasInformedBy and ControlledBy with WasAssociatedWith. PROV adds a recursive dependency for the Agent type as well as a dependency of Entity on Agent.
In summary, we showed (not strictly formally) that all reviewed base models used for medical applications can be represented in terms of the base PROV model. The models are then part of a model lineage leading from the OPM to PROV, with transitional models in between.
Since the reviewed models were designed or extended for and applied in actual biomedical applications we conclude that there exists general and standardised provenance models for this use case. This does not necessarily mean that all biomedical use cases are covered by PROV, the most general model we found. Even if its extensibility was shown on a use case basis in the reviewed works, each new and specific use case should perform its own requirements analysis and – preferably also quantitative – evaluation.
Discussion
When looking at the figures shown in the raw-data and summary tables the likely impression is one of heterogeneity, apart from the directed acyclic graph that underlies all but one model representations. Differences are in the specific application, (visual) structure and even terminology used to describe the models.
However, this impression of heterogeneity proves misleading on further examination. A closer look already reveals elements that are shared by most models: entities, agents (or actors), activities and the relations between them. This similarity is not accidental. Two of the modelling articles base their work on the Open Provenance Model (OPM) : and . The OPM was first released in 2007 and served as a basis for developing .
and also appear to lead up to the development of the PROV data model. Not only do they cover aspects of said model, whose proposed documentation status in 2013 they precede, but the intersection of authors of both papers are all contributors to the W3C model.
Next, and directly use and extend the PROV data model.
It emerges that six out of the seven provenance modelling articles considered in the review base their models on the same lineage of models – which are all part of the provenance of PROV. The remaining work is , which aims to create a model instance for a particular purpose and does not use the existing OPM or PROV data model but can be represented using PROV as shown above.
Since most models share a common basis, there are also similarities in the development process. Mostly, model design is informed by a requirements analysis, often in the form of use cases (also called provenance questions), and evaluation is done with respect to those requirements, albeit largely in a qualitative fashion that shows little more than the model’s feasibility.
Summary
In this scoping review, we captured works which examine the modelling of data provenance in the context of biomedical applications. While provenance modelling, tracking, querying and other provenance applications are all within the review’s initial scope, we confined further analysis in this work to the references dealing primarily with provenance modelling aspects. To our knowledge this is the first review using systematic literature retrieval methods on this topic, which aim to minimise the risk of bias.
Despite their heterogeneous presentation involving non-standard diagrams and different levels of abstraction we could reduce all found base models for biomedical applications to the PROV data model.
Our result follows from traceable, systematic search, simplification, and standardisation steps. We depicted the biomedical provenance models found using systematic literature search using a standard representation, simplified and abstracted where possible, without changing the underlying base model. The final representation enables comparisons that would hardly be comprehensible using the original data extracted from the reviewed articles.